1
Anti-SBPase | Sedoheptulose-1,7-bis phosphatase
AS15 2873 | Clonality: Polyclonal | Host: Rabbit | Reactivity: A. thaliana, Chlamydomonas sp. (strain W80), C. sativus, G. hirsutum, H. vulgare, O. sativa, Z. mays
Benefits of using this antibody
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide conserved across known protein sequences of SBP including Arabidopsis thaliana UniProt: P46283, TAIR: AT3G55800
Host: Rabbit Clonality: Polyclonal Purity: Immunogen affinity purified serum in PBS pH 7.4. Format: Lyophilized Quantity: 50 µg Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Western blot (WB) Recommended dilution: 1 : 1000-1 : 5000 (WB) Expected | apparent MW: 42 kDa
- Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Chlamydomonas sp. (strain W80), Cucumis sativus, Gossypium hirsutum, Hordeum vulgare, Oryza sativa, Setaria viridis, Zea mays Predicted reactivity: Arabidopsis lyrata, Auxenochlorella protothecoides, Brachypodium distachyon , Brassica rapa, Coccomyxa subellipsoidea C-169 GN, Dunaliella tertiolecta, Ectocarpus siliculosus, Genlisea aurea, Glycine soja, Gossypium raimondii , Marchantia polymorpha, Medicago truncatula, Mesostigma viride, Morus notabilis, Nannochloropsis sp., Nicotiana tabacum, Populus trichocarpa, Ricinus communis, Sorghum bicolor, Spinacia oleracea, Tetraselmis sp. GSL018, Theobroma cacao, Triticum aestivum, Triticum urartu, Zea mays, Volvox carteri
Species of your interest not listed? Contact usNot reactive in: Cyanobacteria, Physcomitrella patens - Application Examples
-

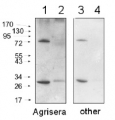
10 µg of total protein from Arabidopsis thaliana leaf (1) , SBPase protein standard AS15 2873S (2) were extracted with Agrisera Protein Extraction Buffer PEB (AS08 300). Samples were diluted with 1X sample buffer (NuPAGE LDS sample buffer (Invitrogen) supplemented with 50 mM DTT and heat at 70°C for 5 min and keept on ice before loading. Protein samples were separated on 4-12% Bolt Plus gels, LDS-PAGE and blotted for 70 minutes to PVDF using tank transfer. Blots were blocked immediately following transfer in 2% blocking reagent or 5% non-fat milk dissolved in 20 mM Tris, 137 mM sodium chloride pH 7.6 with 0.1% (v/v) Tween-20 (TBS-T) for 1h at room temperature with agitation. Blots were incubated in the primary antibody at a dilution of 1: 10 000 (in blocking reagent) for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, and then washed 1x15 min and 3x5 min with TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated, recommended secondary antibody AS09 602, Agrisera) diluted to 1:25 000 in blocking reagent for 1h at room temperature with agitation. The blots were washed as above. The blot was developed for 5 min with chemiluminescent detection reagent of extreme femtogram sensitivity, according the manufacturers instructions. Images of the blots were obtained using a CCD imager (VersaDoc MP 4000) and Quantity One software (Bio-Rad). Exposure time was 30 seconds.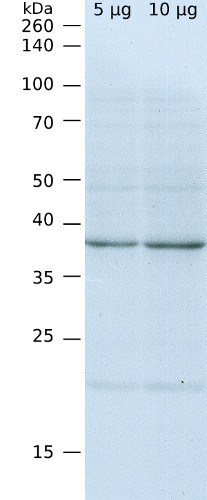
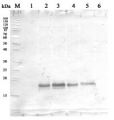
5 or 10 µg of total protein from Chlamydomonas reinhardtii extracted with 10 mM Tris/HCl pH7.5, 80 mM NaCl, 1 mM EDTA, 1 % (w/v) Glycerol, 1 mM DTT, 1x protease inhibitor cocktail (Roche) and denatured in SDS sample buffer for 5 min. at 95°C were separated on 12 % SDS-PAGE and blotted over night to nitrocellulose using tank transfer. Blots were blocked with 3 % milk; for 1h at room temperature (RT) with agitation. Blot was incubated in the primary antibody at a dilution of 1: 2 500 in blocking buffer for 1h at RT with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 5 min. 5x in TBS-T (0.05 %) at RT with agitation. Blot was incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated from Agrisera AS09 602) diluted to 1:25 000; for 1h at RT with agitation. The blot was washed as above and developed using Luminol-H2O2 and p-Coumaric acid. Exposure time of X-ray film was 1 hour.
Courtesy of Dr. Sebastian Mahlow, Prof. Maria Mittag, Institute of General Botany and Plant Physiology, University of Jena, GermanyApplication examples: 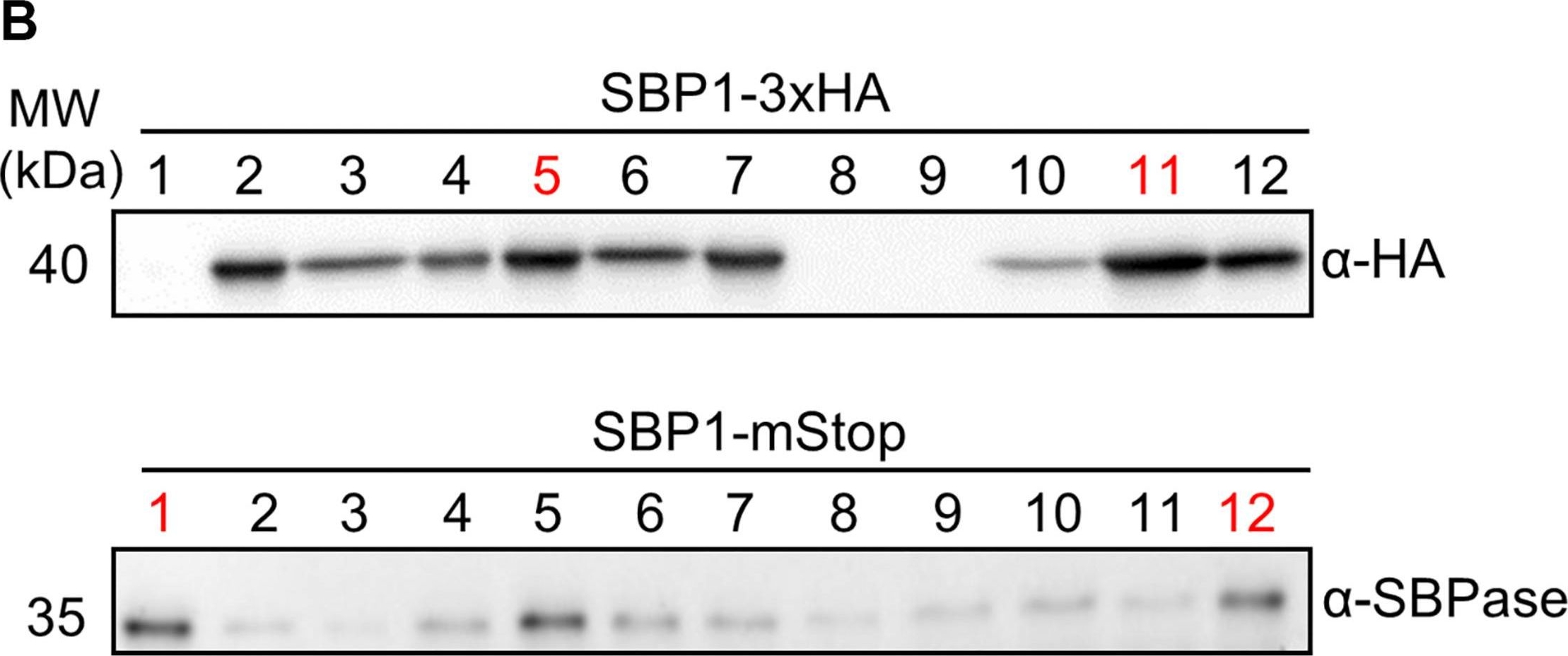
Reactant: Chlamydomonas reinhardtii (Green Alga)
Application: Western Blotting
Pudmed ID: 32655601
Journal: Front Plant Sci
Figure Number: 1B
Published Date: 2020-07-14
First Author: Hammel, A., Sommer, F., et al.
Impact Factor: 5.435
Open PublicationGeneration of Chlamydomonas lines overexpressing SBP1. (A) SBP1 construct used for transformation. The 2172 bp SBP1 ORF (exons shown as black boxes), interrupted by all native SBP1 introns (thin lines), was domesticated to generate a level 0 module for the MoClo strategy. Using MoClo, the SBP1 ORF was equipped with the HSP70A-RBCS2 promotor and the RPL23 terminator (pAR and tRPL23, respectively, white boxes) and with a 3xHA-tag or without a tag (mStop) (gray box), giving rise to two level 1 constructs. These were combined with another level 1 construct containing the aadA gene conferring resistance to spectinomycin (light gray box) to yield the final level 2 constructs for transformation. (B) Screening of transformants overexpressing SBP1. The UVM4 strain was transformed with the level 2 constructs shown in (A). Total cell proteins from 12 spectinomycin resistant transformants recovered with each construct were extracted and proteins corresponding to 1.5 ?g chlorophyll were analyzed by immunoblotting using anti-HA or anti-SBPase antibodies. The transformant number is given on top of the lanes. Transformants exhibiting highest expression levels for SBP1-3HA or SBP1-mStop (red) were used for further analysis.
- Background
-
Background: SBPase (Sedoheptulose-1,7-bis phosphatase) is a chloroplast enzyme involved in the carbon reduction of the Calvin cycle, part of carbohydrate metabolism. - Product Citations
-
Selected references: Ermakova et al. (2024). Chloroplast NADH dehydrogenase-like complex-mediated cyclic electron flow is the main electron transport route in C4 bundle sheath cells. New Phytol. 2024 Jul 22.doi: 10.1111/nph.19982.
Li et al. (2021) Did breeding alter the light environment, photosynthetic apparatus and photosynthetic capacity of wheat leaves? J Exp Bot. 2021 Nov 10:erab495. doi: 10.1093/jxb/erab495. Epub ahead of print. PMID: 34758079.
Hammel et al. (2020) Photosynthesis in Chlamydomonas reinhardtii and Has No Effect on the Abundance of Other Calvin-Benson Cycle Enzymes. Front Plant Sci. 2020 Jun 23;11:868. doi: 10.3389/fpls.2020.00868. PMID: 32655601; PMCID: PMC7324757.
Fukayama et al. (2018). Expression level of Rubisco activase negatively correlates with Rubisco content in transgenic rice. Photosynth Res. 2018 May 30. doi: 10.1007/s11120-018-0525-9.
Li et al. (2018). Comparative proteomic analysis of key proteins during abscisic acid-hydrogen peroxide-induced adventitious rooting in cucumber (Cucumis sativus L.) under drought stress. Journal of Plant Physiology Volume 229, October 2018, Pages 185-194. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Quantitative Immunoblotting in Plant Sciences using Global Antibodies
Amanda Cockshutt1 & Joanna Porankiewicz-Asplund2
1.Environmental Proteomics NB Inc. Sackville NB CANADA,
2.Agrisera AB, Vännäs, SWEDEN,Summary:
Detecting and quantitating important proteins in plants and other photosynthetic organisms has become more practical and powerful with the application of quantitative immunoblotting using global antibodies which recognize proteins evenly across a wide range of taxa. While many tools are commercially available for the examination of medically relevant proteins in mammals, such tools have been lacking for the study of pressing questions in other organisms. Here we present the advantages of quantitative immunoblotting and the methodology necessary to apply these tools.Introduction | The exquisite specificity of the antibody antigen interaction has long been exploited by protein biochemists. Antibodies bind to small regions on proteins, termed epitopes, which often consist of only a few amino acids. This binding is highly specific
and of high affinity. As each antibody recognizes only a small part of the whole protein, an immunogenic peptide can be selected using sequence alignments. This short peptide is highly conserved and exclusive to the protein under study. The resulting antibodies are then immunoreactive with the conserved peptide region within intact proteins derived from a number of species. We refer to these as Global Antibodies, and they provide both scientific and practical advantages. In non-medical areas such as plant sciences, comparative analyses across species, is often productive; Global Antibodies permit such cross-taxon studies with level detection efficiencies across wide taxonomic ranges. From a practical standpoint, a Global Antibody functioning across a wide taxonomic range reduces the cost of antibody production and stocking by creating a detection tool useful in a wide range of studies.
The ELISA, or Enzyme Linked ImmunoSorbent Assay, employs specific antibodies in a high through put application for the measurement of specific substances in samples of diverse origins. While some sample and antibody combinations can be optimized for detections using ELISA, others are less successful. This can be the result of antibodies with significant cross reactivities, samples that do not adhere optimally to the plate, a lack of quantitated standards or samples of variable quality that display differential reactivity in ELISA. A similar approach is the quantitative immunoblot. As such, it uses the same principles as the ELISA, a reliance on the highly specific antibody antigen interaction to measure the amount of a given protein in a complex mixture. The
major distinguishing feature of this method is the inclusion of a step to electrophoretically separate the proteins in the sample according to their molecular weights. Thus, all samples can be solubilized using a standardized procedure and analyzed concurrently for a number of interesting proteins. The separation step allows the researcher to be more confident that the signal generated actually results from the target protein in question because it is spatially resolved from cross-reactions which would confound an ELISA system.Limitations of Traditional Techniques | A number of traditional techniques are available for the measurement of different functions or complexes of photosynthetic organisms. These include an array of enzyme and functional assays. A disadvantage of such approaches is that they require individually specialized sample harvesting and preparation techniques, and often specialized equipment or reagents for each assay. Therefore a single sample preparation cannot be used to quantify different protein complexes. Furthermore, many samples are not available in the quantities or preservation states necessary for multiple determinations of different protein complexes.
Advantages of Immunodetections with Global Antibodies | When Global Antibodies are used for quantitative immunodetections even small, mixed samples from various harvesting procedures can be assessed for multiple complexes simultaneously. Since all detections employ a common solubilization protocol as a starting point, comparisons between complexes can be made with greater confidence. Since Global Antibodies recognize short highly conserved regions of the protein targets they can be applied to a very broad range of target organisms. They can also be applied with considerable confidence to uncharacterized or mixed samples.
Quantitated recombinant protein standards have been generated to be used as a calibration with Global Antibodies. The target peptide antigen is perfectly conserved in these standard proteins, thus they can be applied for comparative quantitation of the target protein from all species.
Standards and antibodies available
Photosystem I Anti-PsaC (AS04 042) | standard: recombinant PsaC(AS04 042S)
Photosystem II Anti-PsbA C-terminal (AS01 016, AS05 084) | Anti-PsbA N-terminal | (AS06 124) | standard: recombinant PsbA (AS01 016S) | Anti-PsbB (AS04 038) | Anti-PsbD (AS06 146) |
ATP Synthase Anti-AtpB (AS03 030, AS05 085) | standard: recombinant AtpB (AS03 030S) |
RuBisCO Anti-RbcL RbcL (AS01 017, AS03 037)| standard: Rubisco protein (AS03 017S)
Nitrogen Metabolism: Anti-GlnA (AS01 018) | standard: recombinant GlnA (AS01 018S) |
Anti-NifH (AS01 021A) | standard: recombinant NifH (AS01 021S)
Methodology | Sample Preparation. Plant samples are generally ground with liquid nitrogen in a mortar and pestle. The resulting powder is transferred to a plastic tube. Algal samples can be either concentrated by centrifugation or, preferably, by filtration onto glass fiber filters.
Solubilization is performed in Agrisera extraction buffer PEB (AS08 300) , 0.1mg/mL PefaBloc SC (AEBSF) protease inhibitor (Roche). Disruption is most optimally obtained through flash freezing of the sample in liquid nitrogen alternated with thawing by sonication with a microtip. This process can be repeated depending on the toughness of the sample. The sample is adjusted to 50 mM dithiothreitol and heated to 70°C for 5 minutes. Samples are cooled and centrifuged briefly prior to electrophoresis. Optimal quantitation is achieved using moderate sample loads per gel lane, generally 0.5 to 2.5 ug total protein, depending on the abundance of the target protein.
Electrophoresis and Immunoblotting: Once solubilized, the proteins can be separated electrophoretically in a number of systems. We obtain optimal results with the Invitrogen NuPAGE gel system using Bis-Tris 4-12% gradient gels. Proteins are separated in MES SDS running buffer according to the manufacturer’s recommendations at 200 V for 35 minutes. The gels are transferred to PVDF in the same apparatus, the SureLock XCell blot module, for 60 minutes at 30 V for a single gel or 80 minutes for a pair. Following transfer the blots are blocked in 2% blocking agent in tris buffered saline with 0.1% Tween 20 (TBS-T) for 1 hour at room temperature with gentle agitation. The blot is incubated with primary antibody, usually at 1:25 000 to 1:50 000 diluted in 2% blocking agent, for 1 hour at room temperature. For quantitation a relatively high primary antibody:target protein ratio gives more reliable results than immunoblots at low ratios of primary antibody:target protein.
The blot is washed extensively in TBS-T (twice briefly, once for 15 minutes and three times for five minutes). The blot is incubated with secondary antibody, for example goat anti-rabbit IgG horse radish peroxidase conjugated, at 1:50,000 in 2% blocking agent, for 1h/RT with agitation. The blot is washed as above and developed with chemiluminescence detection reagent in mid picogram or extreme femtogram range.Quantitation | When quantitated standards are included on the blot, the samples can be quantitated using the available software. We obtain excellent quantitation with images captured on the Bio-Rad Fluor-S-Max or equivalent instrument using Bio-Rad QuantityOne software. The contour tool is used to select the area for quantitation and the values are background subtracted to give an adjusted volume in counts for each standard and sample. Using this protocol allows us to generate linear standard curves over 1-1.5 orders of magnitude range in target load. It is important to note that immunodetections usually show a strongly sigmoidal signal to load response curve, with a region of trace detection of low loads, a pseudolinear range and a region of saturated response with high loads. For immunoquantitation it is critical that the target proteins in the samples and the standard curve fall within the pseudolinear range. Our total detection range using this protocol spans over 2 orders of magnitude, but the quantifiable range is narrower.
Example | Undergraduate students performed a light shift experiment on two cyanobacterial strains, Synechococcus elongatus PCC 7942 and Anabaena sp. PCC 7120. The students prepared 4 samples for each species and loaded 0.1 ug chlorophyll per lane. Standard lanes of PsbA (D1) protein contained 0.05 pmoles , 0.15 pmoles and 0.45 pmoles. The blots were processed as described above with the chicken anti-PsbA antibody used at 1:20,000 and rabbit anti-chicken IgY-HRP secondary antibodies at 1:50,000. The resulting blot is shown in Figure 1.

Figure 1: An example immunoblot of samples of Synechococcus elongatus PCC 7942 (lanes 1-4) and Anabaena sp. PCC 7120 (lanes 5-8). Molecular weight markers (MagicMark XP, Invitrogen) are in lane 9. Recombinant PsbA protein standards are loaded in lanes 10-12 at 0.05 pmoles, 0.15 pmoles and 0.45 pmoles.

Figure 2: Standard curve for the blot in Figure 1. Picomoles of quantitated standard is plotted on the X-axis versus the adjusted signal volume obtained on the Y-axis.
Conclusion | Quantitative immunoblotting is a powerful but simple tool for the measurement of protein complexes from photosynthetic and other organisms. This technique can be applied to a wide range of species with excellent results, because Global Antibodies raised against conserved peptides can be shared across research on different species and research areas like: ecophysiology, microbial ecology, molecular physiology, proteomics and oceanography.
References | Bouchard J, Suzanne Roy S, Campbell DA (2006) Ultraviolet-B Effects on the Photosystem II-D1 Protein of Phytoplankton Species and Natural Phytoplankton Communities Photochemistry and Photobiology, 82: 936-951
Campbell DA, Cockshutt AM, Porankiewicz-Asplund J (2003) Analyzing Photosynthetic Complexes in Uncharacterized Species or Mixed Phytoplankton Communities using Global Antibodies. Physiologia Plantarum 119: 322-327
MacKenzie TDB, Johnson JM, Cockshutt AM, Burns RA, Campbell DA (2005) Large reallocations of carbon, nitrogen and photosynthetic reductant among phycobilisomes, photosystems and Rubisco during light acclimation in Synechococcus elongatus are constrained in cells under low environmental inorganic carbon. Archives of Microbiology, 183: 190 - 202
Recommended secondary antibodies: goat anti-rabbit HRP conjugated, goat anti-rabbit ALP conjugated
Recommended chemiluminescent detection reagent: AgriseraECLBright
Agrisera Western Blot protocol and video tutorialsQuestions? You are welcome to contact us.
- Reviews:
-
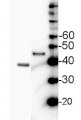
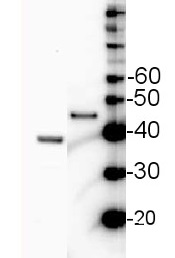
Janna Setzkorn | 2024-02-12Whole cell samples of Chlamydomonas reinhardtii corresponding to 2 µg chlorophyll were separated on 12 % SDS-PAGE (Laemmli-type) and blotted for 1.5 h on a PVDF membrane. After blocking (3% milk in TBS-T) for 1h at RT, blots were incubated in the primary antibody at a dilution of 1:1000 overnight at 4 °C with agitation. The primary antibody, diluted in 3% milk in TBS-T could only be reused once. The secondary HRP-linked anti-rabbit antibody was applied at a dilution of 1:1000 for about 1h at RT. However, using ECL detection, individual bands of SBPase could only be detected in samples from Chlamydomonas lines overexpressing SBPase, but not in the wild-type control.Dorothée Klein | 2023-03-23We analyzed whole cell samples of Chlamydomonas reinhardtii using 10 µg total protein per lane. After WB transfer, we used an AB dilution of 1:3000. The product worked good but I recommend to increase the amount of antibody or the amount of total protein used to get a better signal. We obtained signals for SBPase at approximately 42 kDa.