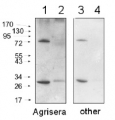
1
RbcL | Rubisco positive control/quantitation standard
AS01 017S | Protein standard for quantitation and positive control, for use in Western blot.
- Product Info
-
Format: Lyophilized in glycerol. Quantity: 100 µl Reconstitution: For reconstitution add 90 µl of sterile water, Please notice that this product contains 10% glycerol and might appear as liquid but is provided lyophilized Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Western blot (WB) Recommended dilution: Standard curve: three protein standard loads are recommended.
For most applications a sample load of 0.2 μg of chlorophyll/well will give a RbcL signal in this range.Positive control: a 2 μl load per well is optimal for most chemiluminescent detection systems. Higher standard concentration needs to be used to allow detection by Coomasie stains (standard concentration 7.9 µg/mL). Such gels with higher standard concentration can not be used for quantitation using chemiluminescence.
This standard is stabilized does not require heating before loading on the gel or addition of any buffer.
Please note that this product contains 10% glycerol and might appear as liquid but is provided lyophilized. Allow the product several minutes to solubilize after adding water. Mix thoroughly but gently Take extra care to mix thoroughly before each use, as the proteins tend to settle with the more dense layer after freezing.Expected | apparent MW: 52.7 kDa - Application Examples
-
Application example

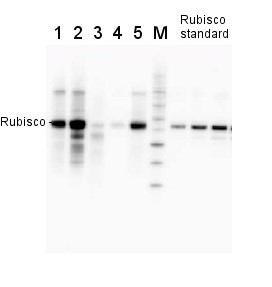
2 µg of total protein from various plant extracts (1-5) extracted with PEB (AS08 300) separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Markers MagicMarks (Invitrogen) (M) and Rubisco protein standard (AS01 017S) at 0.0625 pmol, 0.125 pmol, 0.25 pmol.
Following standard western blot procedure this image has been obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad). The contour tool of the software is used to the area for quantitation and the values are background subtracted to give an adjusted volume in counts for each standard and sample.
Note: Optimal quantitation is achieved using moderate sample loads per gel lane, generally 0.5 to 2.5 ug total protein, depending on the abundance of the target protein.Quantitation: When quantitated standards are included on the blot, the samples can be quantitated using the available software. Excellent quantitation can be obtained with images captured on the Bio-Rad Fluor-S-Max or equivalent instrument using Bio-Rad QuantityOne software. The contour tool is used to the area for quantitation and the values are background subtracted to give an adjusted volume in counts for each standard and sample. Using above protocol linear standard curves are generated over 1-1.5 orders of magnitude range in target load. It is important to note that immunodetections usually show a strongly sigmoidal signal to load response curve, with a region of trace detection of low loads, a pseudolinear range and a region of saturated response with high loads. For immunoquantitation it is critical that the target proteins in the samples and the standard curve fall within the pseudolinear range. Our total detection range using this protocol spans over 2 orders of magnitude, but the quantifiable range is narrower.
Quantitative western blot: detailed method description. - Additional Information
-
Additional information: The RbcL protein standard can be used in a combination with Agrisera global antibiodies (AS01 017 from chicken or AS03 037 from a rabbit) to quantitate RbcL from a wide range of species. Global antibodies are raised against highly conserved amino acid sequence. This standard is also included in following kits: Educational antibody kit - photosynthesis, Photosynthesis Tool Kit - quantitation, Rubico quantitation kit,
Quantitative western blot: detailed method description, video tutorial
Additional information (application): Concentration: after re-constitution with sterile milliQ water final concentration of the standard is 0.15 pmoles/µl
Protein standard buffer composition: Glycerol 10%, Tris Base 141 mM, Tris HCl 106 mM, LDS 2%, EDTA 0.51 mM, SERVA® Blue G250 0.22 mM, Phenol Red 0.175 mM, pH 8.5, 0.1mg/ml PefaBloc protease inhibitor (Roche), 50 mM DTT.
This standard is ready-to-load and does not require any additions or heating. It needs to be fully thawed and thoroughly mixed prior to using. Avoid vigorous vortexing, as buffers contain detergent. Following mixing, briefly pulse in a microcentrifuge to collect material from cap.
This standard is stabilized and ready and does not require heating before loading on the gel.
Please note that this product contains 10% glycerol and might appear as liquid but is provided lyophilized. Allow the product several minutes to solubilize after adding water. Mix thoroughly but gently Take extra care to mix thoroughly before each use, as the proteins tend to settle with the more dense layer after freezing.Please, use the 55 kDa size of RbcL for calculations. The pmoles in the standard refer to pmoles of rbcL monomers.
Why can I not see the standard band using Coomasie stain? The reason that you do not see Rubisco standard on a gel is, that you have probably used it in concentration which is recommended for western blot detection, and it is too low to allow to see this protein using Coomasie stain. In such a case, you should load more Rubisco standard on a gel and stain it with more sensitive Coomasie stain or with silver. You can not use such a gel for western blot, as using higher concentration of this standard will not work for quantitation using western blot technique.
- Background
-
Background: Rubisco (Ribulose-1,5-bisphosphate carboxylase/oxygenase) catalyzes the rate-limiting step of CO2 fixation in photosynthesis. It is one of the most abundant proteins on Earth and its homology has been demonstrated from purple bacteria to flowering plants.
Source of Rubisco standard: Rubisco protein was purified directly from a plant tissue - spinach.
- Product Citations
-
Selected references: Aguiló-Nicolau et al. (2023) Singular adaptations in the carbon assimilation mechanism of the polyextremophile cyanobacterium Chroococcidiopsis thermalis. Photosynth Res. 2023 May;156(2):231-245. doi: 10.1007/s11120-023-01008-y.
Capo-Bauca et al. (2023). Carbon assimilation in upper subtidal macroalgae is determined by an inverse correlation between Rubisco carboxylation efficiency and CO2 concentrating mechanism effectiveness. New Phytol. 2023;237(6):2027-2038. doi:10.1111/nph.18624
Capo-Bauca et al. (2022) Correlative adaptation between Rubisco and CO2-concentrating mechanisms in seagrasses. Nat Plants. 2022 Jun;8(6):706-716. doi: 10.1038/s41477-022-01171-5. Epub 2022 Jun 20. Erratum in: Nat Plants. 2022 Jun 29;: PMID: 35729266.
Perera-Castro et al (2022). Limitations to photosynthesis in bryophytes: certainties and uncertainties regarding methodology. J Exp Bot. 2022;73(13):4592-4604. doi:10.1093/jxb/erac189
Poor et al. (2018). Comparison of changes in water status and photosynthetic parameters in wild type and abscisic acid-deficient sitiens mutant of tomato (Solanum lycopersicum cv. Rheinlands Ruhm) exposed to sublethal and lethal salt stress. J Plant Physiol. 2018 Dec 8;232:130-140. doi: 10.1016/j.jplph.2018.11.015.
Dai et al. (2018). Visualizing Individual RuBisCO and its Assembly into Carboxysomes in Marine Cyanobacteria by Cryo-Electron Tomography. J Mol Biol. 2018 Aug 20. pii: S0022-2836(18)30411-X. doi: 10.1016/j.jmb.2018.08.013.
Li et al. (2016). Interactive effects of nitrogen and light on growth rates and RUBISCO content of small and large centric diatoms. Photosynth Res. 2016 Aug 26. [Epub ahead of print].
Young et al. (2015). Antarctic phytoplankton down-regulate their carbon-concentrating mechanisms under high CO2 with no change in growth rates. Marine Ecology Progress Series 532:13-28.
Vandenhecke et al. (2015). Changes in the Rubisco to photosystem ratio dominates photoacclimation across phytoplankton taxa. Photosynth Res. 2015 Apr 11.
Wu et al. (2014). Large centric diatoms allocate more cellular nitrogen to photosynthesis to counter slower RUBISCO turnover rates. Front. Mar. Sci., 09 December 2014 | doi: 10.3389/fmars.2014.00068.
Li et al. (2014). The nitrogen costs of photosynthesis in a diatom under current and future pCO2. New Phytol. 2014 Sep 25. doi: 10.1111/nph.13037. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extractsRubisco quantitation in plant and algal samples using Agrisera anti-RbcL global antibody and Rubisco protein standard
Methodology: Plant samples are generally ground with liquid nitrogen in a mortar and pestle. The resulting powder is transferred to a plastic tube. Algal samples can be either concentrated by centrifugation or, preferably, by filtration onto glass fiber filters. Solubilization is performed in Agrisera protein extraction buffer (PEB, AS08 300) containing 0.1mg/mL PefaBloc SC (AEBSF) protease inhibitor (Roche). Disruption is most optimally obtained through flash freezing of the sample in liquid nitrogen alternated with thawing by sonication with a microtip. This process can be repeated depending on the toughness of the sample. The sample is adjusted to 50 mM dithiothreitol and heated to 70°C for 5 minutes. Samples are cooled and centrifuged briefly prior to electrophoresis.
Optimal quantitation is achieved using moderate sample loads per gel lane, generally 0.5 to 2.5 ug total protein, depending on the abundance of the target protein.
Electrophoresis and Immunoblotting: Once solubilized, the proteins can be separated electrophoretically in a number of systems. We obtain optimal results with the Invitrogen NuPAGE gel system using Bis-Tris 4-12% gradient gels. Proteins are separated in MES SDS running buffer according to the manufacturer’s recommendations at 200 V for 35 minutes. The gels are transferred to PVDF in the same apparatus, the SureLock XCell blot module, for 60 minutes at 30 V for a single gel or 80 minutes for a pair. Following transfer the blots are blocked with non-fat dry milk up to 10 % in TBS-T, for 1 h/RT with gentle agitation. The blot is incubated with primary antibody, usually at 1:25 000 to 1:50 000, for 1 h/RT (if extreme femtogram detection reagents are used) or in lower primary antibody dilution for less sensitivie reagents (mid picogram and lower).
For quantitation a relatively high primary antibody: target protein ratio gives more reliable results than immunoblots at low ratios of primary antibody:target protein.
The blot is washed extensively in TBS-T (twice briefly, once for 15 minutes and three times for five minutes). The blot is incubated with secondary antibody, for example goat anti-rabbit IgG horse radish peroxidase conjugated, AS09 602 (Agrisera), for 1h/RT. The blot is washed as above and developed with ECL detection reagents.Quantitation: When quantitated standards are included on the blot, the samples can be quantitated using the available software. Excellent quantitation can be obtained with images captured on the Bio-Rad Fluor-S-Max or equivalent instrument using Bio-Rad QuantityOne software. The contour tool is used to select the area for quantitation and the values are background subtracted to give an adjusted volume in counts for each standard and sample. Using above protocol linear standard curves are generated over 1-1.5 orders of magnitude range in target load. It is important to note that immunodetections usually show a strongly sigmoidal signal to load response curve, with a region of trace detection of low loads, a pseudolinear range and a region of saturated response with high loads. For immunoquantitation it is critical that the target proteins in the samples and the standard curve fall within the pseudolinear range. Our total detection range using this protocol spans over 2 orders of magnitude, but the quantifiable range is narrower.
References:
MacKenzie et al (2005). Large reallocations of carbon, nitrogen and photosynthetic reductant among phycobilisomes, photosystems and Rubisco during light acclimation in Synechococcus elongatus are constrained in cells under low environmental inorganic carbon. Arch of Microbiol. 183: 190 - 202.
Bouchard et al. (2006) UVB effects on the photosystem II-D1 protein of phytoplankton and natural phytoplankton communities. Photochem and Photobiol 82: 936-951.
Morash et al. (2007) Macromolecular dynamics of the photosynthetic system over a seasonal developmental progression in Spartina alterniflora. Can J. of Bot. 85: 476-483(8)Application example
.jpg)
2 µg of total protein from (1-7) Prochlorococcus sp. extracted with Protein Extration Buffer, PEB (AS08 300), (8-11) Rubisco protein standard (AS01 017S) were separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Blots were blocked immediately following transfer for 1h/RT with agitation. Blots were incubated in the primary antibody at a dilution of 1: 20 000 for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated) diluted to 1:50 000 for 1h/RT with agitation. The blots were washed as above and developed for 5 min with ECL detection reagent according the manufacturers instructions. Images of the blots were obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad). There is a low level of a known RbcL cleavage product below the main band in some of the Prochlorococcus samples.
Recommended secondary antibodies: goat anti-rabbit HRP conjugated, goat anti-rabbit ALP conjugated
Recommended chemiluminescent detection reagent: AgriseraECLBright - Reviews:
-
Veronika Z | 2014-09-29The standart were used in according with datasheet. Standart separeted in gel and transferred to a polyvinylide fluoride membrane clearly. It was convenient to generate a standard curve and determine the absolute protein concentration of each sample.
Accessories
AS01 017 | Clonality: Polyclonal | Host: Hen | Reactivity: [global antibody] for higher plants, algae, cyanobacteria | cellular [compartment marker] of plastid stroma
Benefits of using this antibody
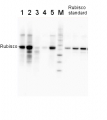
This set enables quantification of Rubisco using Western blot in samples from a range of various species including dicots, monocots, algae, cyanobacteria. Benefits of using this antibody set