
Western blot
Western Blot Troubleshooting Guide

Western Blot Resources
Quantitative western blot video tutorial • Antibody production and validation webinar • Antibody storage guidelines
Secondary antibody collection • Secondary antibody guide • Catalog antibody FAQ
Antibody production FAQ • Agrisera's YouTube channel • Agrisera's technical blog
Western Blot Recommendations and Troubleshooting
Each antibody-antigen interaction has unique characteristics. A protocol giving good results for one antibody-antigen pair might be found to be unsatisfactory for another antibody-antigen pair, even within the same sample. The interaction of antisera with protein epitopes in Western blotting depends on a number of factors, all contributing to the final signal/noise ratio. Knowledge and control of those factors allow for modifications and optimization of the procedure. Polyclonal antibodies (serum or IgY-fractions from egg yolk) consists of a pool of different antibodies, interacting independently with different epitopes (consisting of 3-15 amino acids) on the target used for given antibody production. This number is limited for peptide-antigens, but if recombinant or native proteins have been used for immunization, the signal obtained has to be regarded as cumulative (i.e. caused by several different types of antibodies present in the serum/IgY used).To achieve reproducible results, we recommend the use of gloves, forceps, clean and detergent-free laboratory materials, reagents which have not expired, and freshly prepared buffers with controlled pH. Additionally, it is important to keep protocol volumes, dilutions, and incubation times the same. In general, incubation times should be as short as possible, while primary and secondary antibody dilutions should be as high as possible.
Agrisera has prepared a poster that covers western blot troubleshooting, found here. If interested, you can also request a printed copy.
1. Sample
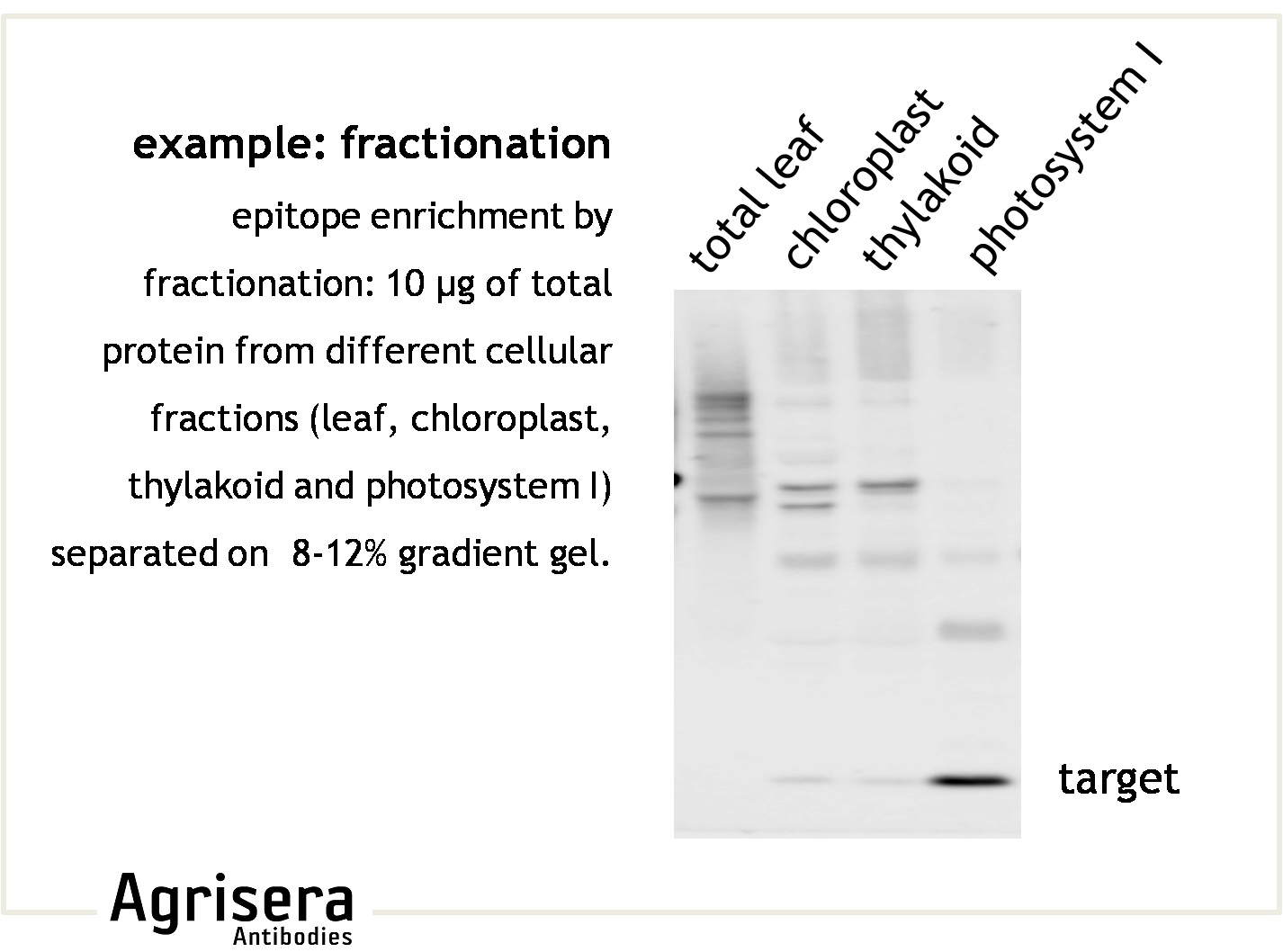
To ensure that the target protein is present in the starting material, it is essential to prepare and store the samples under non-degrading/aggregating conditions. If the target protein is degraded or not present in sufficient amounts in the loaded sample, it can result in lack of signal detection. Epitope abundance can be enhanced under altered growth conditions, by selective tissue preparation, or fractionations of complex cellular extracts (i.e. organelle preparation). Using an extraction method that provides good yields and consistent results is highly recommended, and the method can vary depending on organism and tissue type. Agrisera has validated the Precellys® device from Bertin Technologies, as it fulfills these criteria. For work with organisms that possess robust cell walls, like diatoms, some recommendations can be found here. Useful reference: "What is the total number of protein molecules per cell volume? A call to rethink some published values." Milo, 2013.
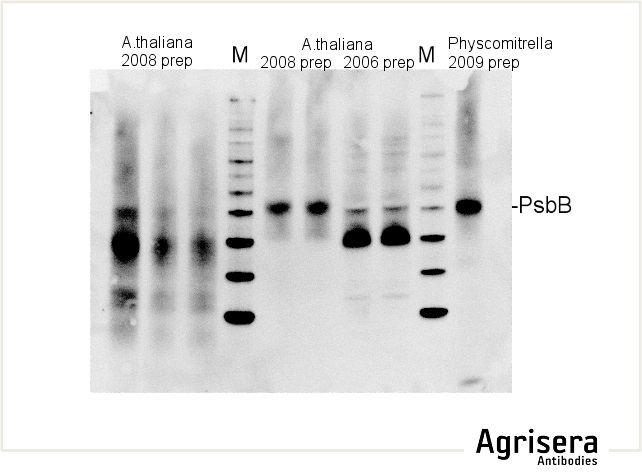
Sample quality is of crucial importance. As shown in the example below, samples of Arabidopsis thaliana total leaf preparations from year 2008 or 2006 show greater degradation of PsbB protein as compared to the newer samples from 2009. M denotes molecular weight markers.
The secondary antibody used was Agrisera's high-titer secondary antibody Goat anti-rabbit, HRP-conjugated (1: 50 000) with a chemiluminescent detection system.
Protein samples older than 6 months to 1 year should not be used. In case of proteins of low abundancy, use of fresh samples and protease inhibitors is highly recommended. In some cases, inhibitors of proteasome are necessary.
Run the sample extraction buffer (and loading buffer) in one lane of your gel to check for contribution of your sample extraction buffer to the background signal.
Alternative preparation methods should be considered if possible.
Include positive and negative controls from the beginning. Comparison of samples prepared from material containing no target (e.g. genetic 0-mutants) or higher amounts of the target (e.g. over-expression mutants) as well as cellular fractionation (e.g. preparation of various fractions of organelles) will allow to control specificity of obtained signals.
Protein extraction protocols: Leaves | Diatoms | Seeds
2. Protein separation
The size-related identification of a protein-antibody interaction usually requires gel separation under fully denaturing conditions. Complete reduction of intra- and intermolecular S-S bridges ensures the accessibility of the epitope for interaction with antibodies detecting linear epitopes. In this case, reducing agents must be added to sufficient final concentrations to both sample and (usually) also the running buffer. If antibodies recognize non-linear epitopes, they require conformational integrity of the target, best provided in non-denaturing PAGE systems or immuno-histochemistry applications. Increasing protein loadings might elevate epitope abundance, but most often also promote non-specific cross-reactions, higher background, and impaired separation.
Avoid protein loadings higher that 5-20 µg/lane for standard mini-gel systems.
Check separation with stained marker pattern or by Coomassie/Silver-staining of the gel. Mixing a pre-stained marker with a marker reacting with the secondary antibody (e.g. MagicMark, Invitrogen) will be an advantage for size-determination and can serve as a control for the visualization assay.
Some highly hydrophobic membrane proteins might require 2-8 M urea in the gels and sample buffer to be kept unfolded during separation or TCA/acetone precipitation method.

3. Protein transfer
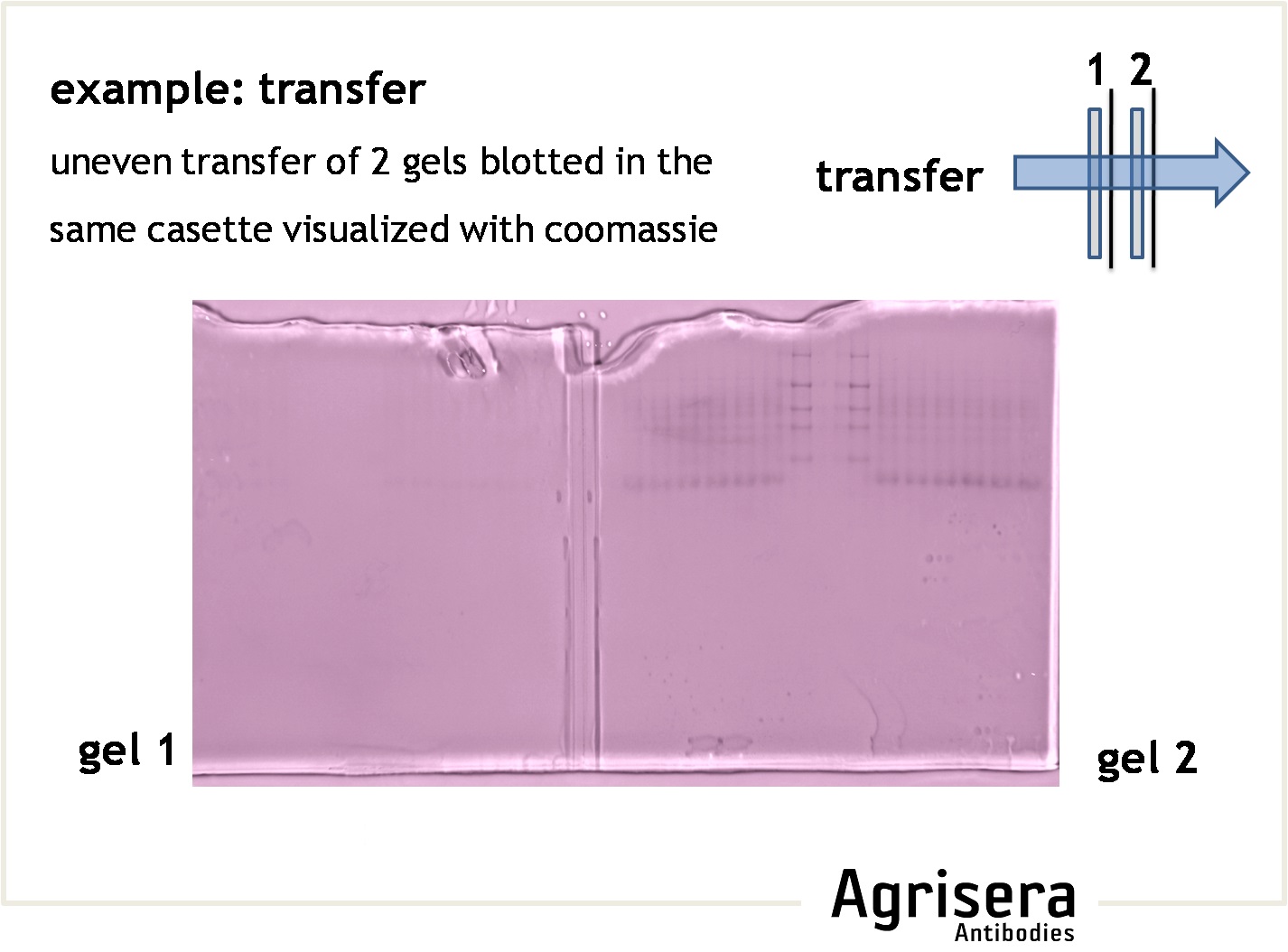
The protein transfer to a suitable membrane (blotting) with protein-binding capacity (nitrocellulose, PVDF) is highly dependent on the biochemical properties of the target protein. Proteins of higher molecular weight (apparent molecular masses of >100 kDa) require longer blotting times than smaller proteins. At low field strengths (<15 V/cm), the mobility of proteins out of the gel will be reduced, and proteins will not be completely transferred from the gel. If field strength is too high (>35 V/cm) proteins might pass through the membrane without binding. The transfer time should be as short as possible to prevent proteins (especially those of lower molecular weight) from passing through the membrane. Place the second membrane and check the signal strength after incubation with primary antibody compare to the first membrane. A short article on protein transfer optimization, prepared by Agrisera staff, can be found here.
- In gradient gels the porosity of the gel is matched with the size of the proteins, that can be an advantage for efficient transfer.
- Check efficiency of a transfer by post-transfer staining of the gel (e.g. Coomassie or Silver) or the filter paper (Ponceau, reversible).
- Drying the membrane (air dry between clean sheets of filter-paper) can improve immobilization of the protein and its denaturation.
- For some antibodies, nitrocellulose membrane might work better compared to PVDF, and for other antibodies it may be the other way around.
- It is crucial that the blotting equipment is well-rinsed with distilled water after each use, and kept away from contaminating detergents. Foam pads should be cleaned and thoroughly washed. When your pads acquire colors (from plant material) and/or loose tension it is time to change them.
- Do not reuse transfer buffers.
- The presence of SDS (0.01 to 0.02 %) in the transfer buffer will increase the mobility of proteins (especially large proteins) out of the gel and provide a negative charge to the protein, which will help to maintain it in its soluble state. At the same time, SDS will reduce protein binding to the membrane (especially nitrocellulose) due to decreased hydrophobicity of the protein.
- The presence of alcohol in the transfer buffer will decrease protein mobility out of the gel. It will also reduce pore size of the gel, while it will improve binding to nitrocellulose as it removes SDS from proteins and increases their hydrophobicity. Higher molecular weight proteins might not be transferred completely if methanol is present in the transfer buffer. Change membrane to nitrocellulose, omit methanol from transfer buffer, while adding SDS and increasing field strength.
- The thickness of the gel will affect protein mobility out of the gel. Thicker gels allow higher loading, but lower molecular weight proteins might transfer less efficiently.
- Hydrophobic proteins might migrate further in the gel than they "should" based on their molecular weight. This is because hydrophobic proteins bind more SDS per amino acid, skewing the charge to mass ration (more negative charge, therefore they migrate faster).
4. Blocking
To minimize background staining due to non-specific membrane-binding of the antibody tested, remaining surfaces of the membrane have to be saturated ("blocked") with proteins that are non-reactive with the antibody. Commonly used are low-fat dry milk powder, casein, IgGs (at 5-10 % but not from the species that your primary antibodies are derived from) or BSA, at 2-5 % w/v. The shortest possible blocking time should be determined experimentally for your system. Increasing the blocking times extensively (e.g. overnight), or incubation at low temperatures, might lead to aggregation of blocking protein on the membrane, and by this compromise later antibody-epitope interactions. If low background is obtained after 30-60 min of blocking, longer blocking will not improve your results. It should be determined experimentally if adding or omitting the blocking reagent in the subsequent antibody incubation steps is an improvement on the results. Normal buffer and detergent concentrations should not reduce the effect of the blocking step, however, accessibility of a target protein for the primary antibody can be increased by a short "block-free" wash. Each antibody-antigen pair is unique and therefore the blocking protocol should be empirically tested.
As some IgY antibodies might recognize milk proteins, BSA might give lower background than milk powder for these antibodies.
Do not use BSA as blocking agent if BSA-coupled peptides were used for immunization. BSA should be of high purity and IgG-free so as not to interfere with the assay.
As milk contains biotin, the use of milk-power for blocking is incompatible with avidin/streptavidin systems.
If serum is used for blocking, care should be taken to ensure that the animal has not been exposed to (or developed antibodies to) the antigen in question. If this is the case, they may bind to the antigen and prevent binding of the primary antibody. Purified immunoglobulins are higher-quality blocking reagents.
As a protein-free protein alternative, you may block for 1 h at RT with 0.2-0.5 % Tween-20 in PBS, followed by incubation with the primary antibodies diluted in 0.25 % Tween-20 in PBS for 1 h at RT.
5. Primary antibody
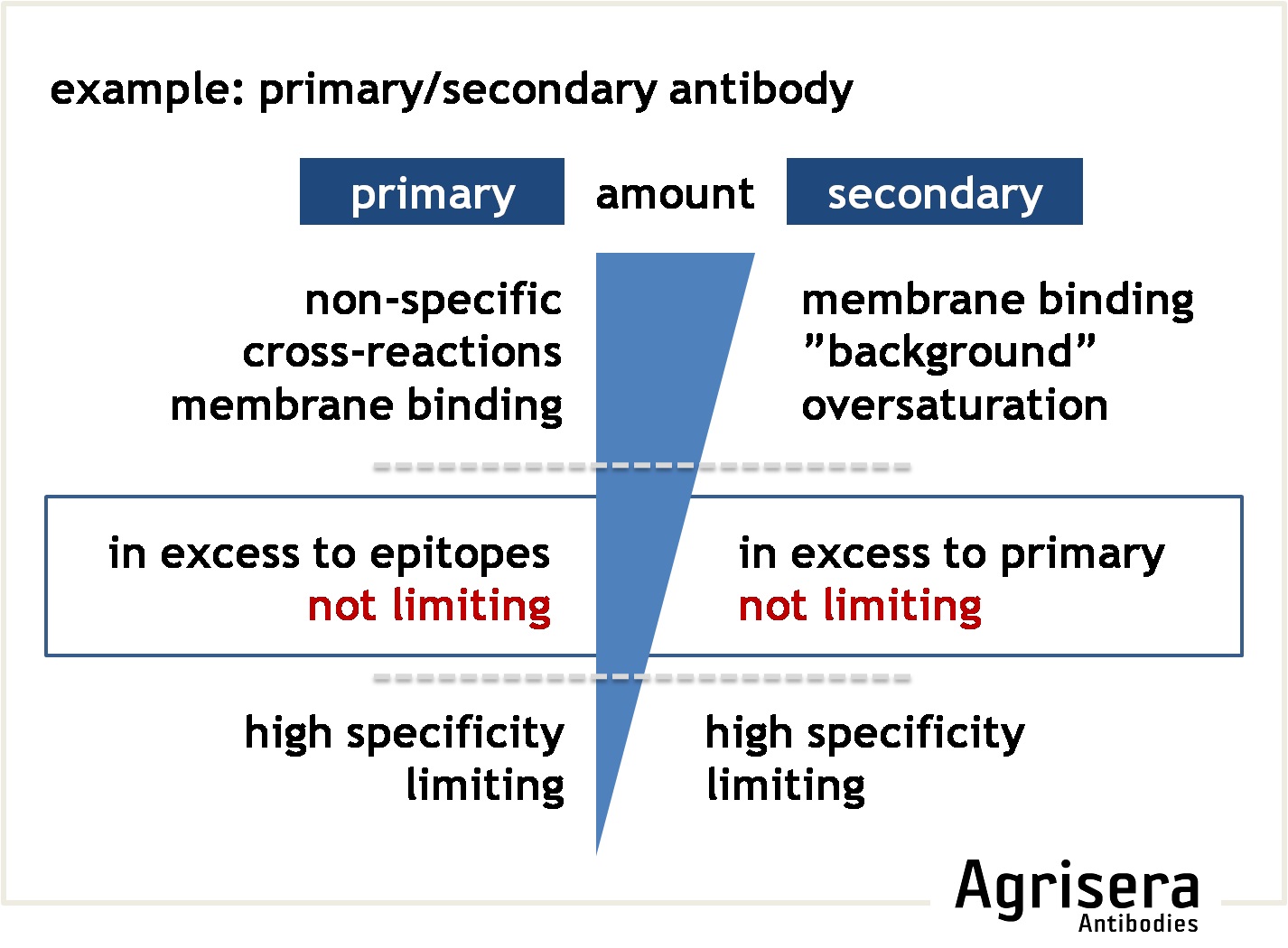
The primary antibody is the major determinant of the specificity of the target-recognition. The interaction with the primary epitope should dominate any cross-reactivity with other epitopes. Commonly used dilutions are between 1:500 to 1:20 000, depending on the reactivity of the antibody and the detection system used. To check for specificity of the target recognition you can (1) use another antibody against your target which will bind to other epitopes on a target protein, (2) run control samples free or depleted of target-proteins, or (3) perform a peptide competition assay (for anti-peptide antibodies).
If available, a second antibody known to react with the sample can be used as a control of subsequent assay steps, on a parallel filter in the same experiment.
Lyophilized primary antibodies incubated for 2-4 h at 4ºC after reconstitution, prior to use, may increase signal strength.
To diminish unspecific cross-reactions, you may pre-adsorb the primary antibody (overnight, 4ºC) with tissue extract lacking your protein of interest (you can use a transferred membrane where the band/area containing your target protein has been cut out). In case of increased background signals, incubate your membrane before any protein is transferred in saturated PBS-milk, followed by protein transfer.
Antibodies against carrier proteins (KLH, BSA or others) might recognize epitopes of other proteins present in the extract. Those anti-carrier antibodies can be removed by (1) incubating serum/IgY sample with a carrier protein in solution (0.1 % (w/v), or spotted on PVDF membrane, or (2) passing serum/IgY sample through the column with bound carrier protein (use flow-through fraction in further experiments), or by affinity purification of the primary antibodies on a column with coupled antigen.
Primary antibodies stored (at 4°C or -20°C) in solution with blocking reagent often lose their reactivity. Antibody dilutions should be stored in buffer only, not buffer + blocking mixture. Re-using of primary antibodies is not recommended as it can contribute to decrease of detected signal, depending on antibody titer.
Lots of small spots visible on a membrane during development can be due to fat in a given serum. To improve such blots, use milk-based blocking reagents as well as increasing Tween concentration in your buffers.
6. Secondary antibody
The secondary antibody has to be reactive against the primary antibody (e.g. use anti-rabbit to detect primary antibodies raised in rabbit), and is usually coupled to an enzyme or dye that allows subsequent visualization. Thus, any non-target binding of the secondary antibody will result in background (if bound to the membrane due to insufficient blocking), or false-positive recognition of non-target proteins present on the filter ("cross-reactions"). Secondary antibodies are usually used at dilutions of 1:20 000-1:500 000, depending on the sensitivity of the visualization method (e.g. chemiluminescence or chromogenic detections). The optimal dilution of the secondary antibody must be determined experimentally for the detection system used. Different secondary antibodies may even result in different recognition patterns when applied to the same sample.
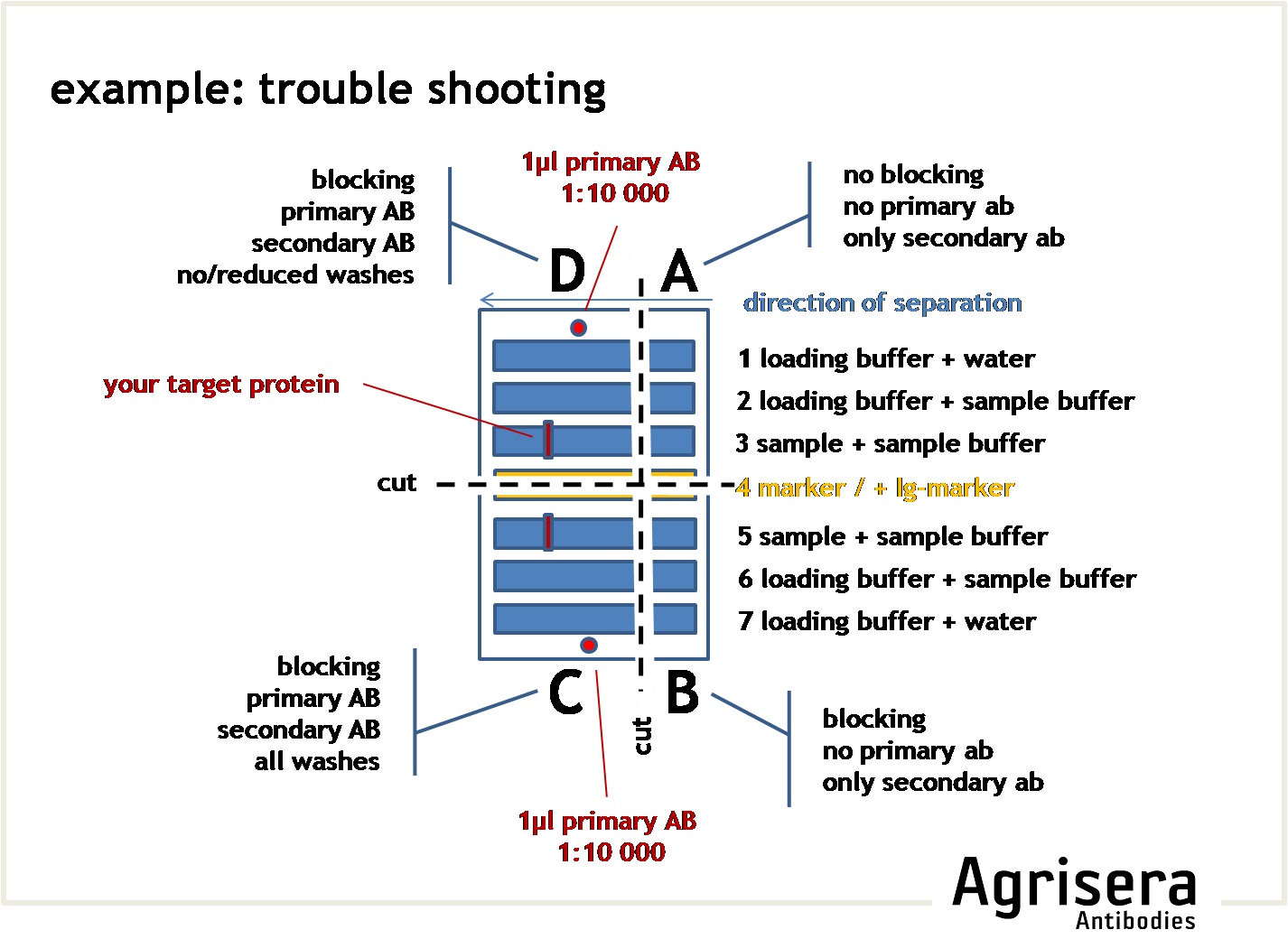
To check the contribution of the secondary antibody on the results, you may cut a suitable square of your filter and run it as a parallel control where the primary antibody is omitted (see example for troubleshooting).
If an Ig-reactive marker has been used, the signals obtained from the marker can serve as a control of the function of the secondary antibody, as well as for size determination.
The sensitivity of the secondary antibodies can differ between various manufacturers.
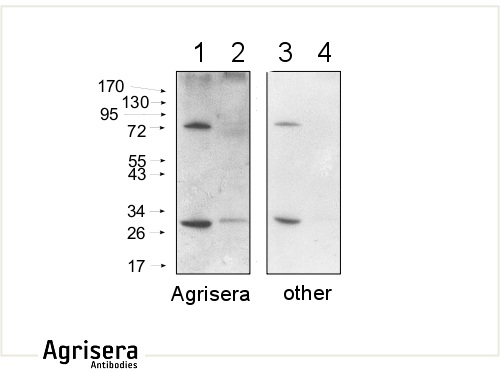
10 μg of mitochondrial fraction from Arabidopsis thaliana (1,3) and Arabidopsis thaliana leaf extract (2,4) were separated on 10 % gel and blotted on nitrocellulose membrane using wet transfer (0.22 % CAPS, pH 11). Filters where blocked (1.5 h) in 5 % milk in TBST (1X TBS, 0.1% Tween 20), incubated with 1: 1000 anti-COXII antibodies (2 h in TBST) followed by incubation with 1: 10 000 secondary anti-rabbit (1 h) HRP-coupled antibodies from Agrisera (left panel) and other manufacture (right panel) and visualized with standard ECL on Kodak autoradiography film for 5 s. The antibody in the left panel detects target protein also in total cell extract (2) and can be used in higher dilution than applied 1: 10 000. Agrisera's goat anti-rabbit HRP conjugated antibody (AS09 602) can be used at following dilutions: 1: 50 000 -1: 90 000 (ELISA), 1 : 75 000 with enhanced ECL and 1: 25 000 with chemiluminescence (WB), 1: 500 -1: 5000 (IHC).
7. Washes
After steps 4, 5 and 6, excess amounts of blocking reagent or antibody need to be diminished by washing the membrane in buffer, usually the same buffer used in the blocking or incubating solutions. Typically, the buffer will include a detergent, such as 0.05 % - 0.5 % of Tween20. A Blocking protein in a 1:10 dilution can be added to the wash buffer to help minimize background. In some protocols, the multiple wash steps omit the detergent in the last step. For reasons of reproducibility, it is recommended to keep volumes and times constant. The intensity of the washing steps can be elevated by (a) increased times and volumes, (b) additional changes of buffer volume, (c) higher detergent concentrations, and (d) use of stronger detergents (e.g. SDS instead of Tween-20). Use high-purity detergents, as they might otherwise contain high amounts of peroxides and contribute to increased background. It is good practice to verify that stocks of buffers (including the wash buffer) do not contain microbial growth (visible as cloudiness that settles), as this can contribute to increased background noise.
8. Detection
Enzymatic detection systems are the most popular ones, and employ usage of secondary antibodies conjugated with Alkaline phosphatase (AP or ALP) or horseradish peroxidase (HRP). However, an alkaline phosphatase detection system is at least 10x less sensitive than a chemiluminescence based system, like ECL. Therefore, using ALP is not recommended when working with low expression level proteins in the tissue. In such cases, more sensitive detection systems should be employed. Agrisera offers a set of two chemiluminescent detection reagents with mid picogram (AgriseraECL Bright) and extreme low femtogram detection range (AgriseraECL SuperBright) combined into a set of two. Some laboratories use fluorophore-conjugated secondary antibodies, such as DyLight®, and in such cases there is no substrate development step in the assay. The protocol is shorter, and due to advances of digital imaging, multiple fluorophores can be used in the same assay.
9. Membrane storage
After western blotting has been completed, the membranes (nitrocellulose or PVDF) can be stored for up to 6 months at room temperature, between sheets of tissue paper (like Kleenex or KimWipes).
