1
Anti-PsaC | PSI-C core subunit of photosystem I
AS10 939 | Clonality: Polyclonal | Host: Rabbit | Reactivity: [global antibody] for higher plants, algae, cyanobacteria, diatoms
Benefits of using this antibody
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide conserved in all known PsaC proteins including Arabidopsis thaliana Uniprot:P62090 TAIR: AtCg01060, Hordeum vulgare UniProt: P69416, Oryza sativa UniProt: P0C360, Chlamydomonas reinhardtii UniProt: Q00914, Synechococcus elongatus UniProt: Q31QV2
Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Western blot (WB) Recommended dilution: 1 : 5000 (WB) Expected | apparent MW: 9 kDa - Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Chlamydomonas reinhardtii, Cyanophora paradoxa, Dactylis glomeRata, Emiliania huxleyi, Euglena gracilis, Gonyaulax polyedra, Horderum vulgare, Lolium perenne, Marchantia polymorpha, Mesembryanthemum sp., Spinacia oleracea, Flaveria sp., Heterosigma akashiwo,Micromonas pusilla, Phaeodactylum tricornutum, Porphyra sp., symbiotic dinoflagellates of Stylophora pistillata and Turbinaria reniformis; Synechococcus sp.PCC7002, sp. PCC 7942, Synechocystis sp. PCC 6803, Thalassiosira pseudonana, Thalassiosira punctigera, Trichodesmium erythraeum, Triticum aestivum Predicted reactivity: Algae, Cannabis sativa, Chromera velia, Cyanobacteria, Brassica napus, Glycine max, Manihot esculenta, Nannochloropsis sp., Nicotiana benthamiana, Nicotiana tabacum, Physcomitrium patens, Prochlorococcus sp. (surface and a deep water ecotype), Spinacia oleracea, Synechococcus PCC 8801, Thermosynechococcus elongatus (BP-1)
Species of your interest not listed? Contact usNot reactive in: No confirmed exceptions from predicted reactivity are currently known - Application Examples
-

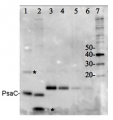
5 µg of total protein from samples such as (1) Arabidopsis thaliana leaf, (2) Hordeum vulgare leaf, (3) Chlamydomonas reinhardtii total cell, (4) Synechococcus sp. 7942 total cell, (5) PsaC protein standard (AS04 042S), total protein from all the samples were extracted with Agrisera Protein Extraction Buffer PEB (AS08 300). Samples were diluted with 1X sample buffer (NuPAGE LDS sample buffer (Invitrogen) supplemented with 50 mM DTT and heat at 70°C for 5 min and kept on ice before loading. Protein samples were separated on NuPAGE 4-12% Tris-Bis gel (Invitrogen) LDS-PAGE and blotted for 1h to 1.5h on PVDF using tank transfer. Blots were blocked immediately following transfer in 2% blocking reagent for 1h at RT with agitation. Blots were incubated with PsaC antibody at a dilution of 1: 10 000 (in blocking reagent) for 1h at RT with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, and then washed 1x15 min and 3x5 min with TBS-T at RT with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG HRP conjugated, AS09 602) diluted to 1:50 000 in blocking reagent for 1h at RT with agitation. The blots were washed as above. The blot was developed for 5 min with chemiluminescence detection reagent in mid picogram range. Images of the blots were obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad).Application examples: 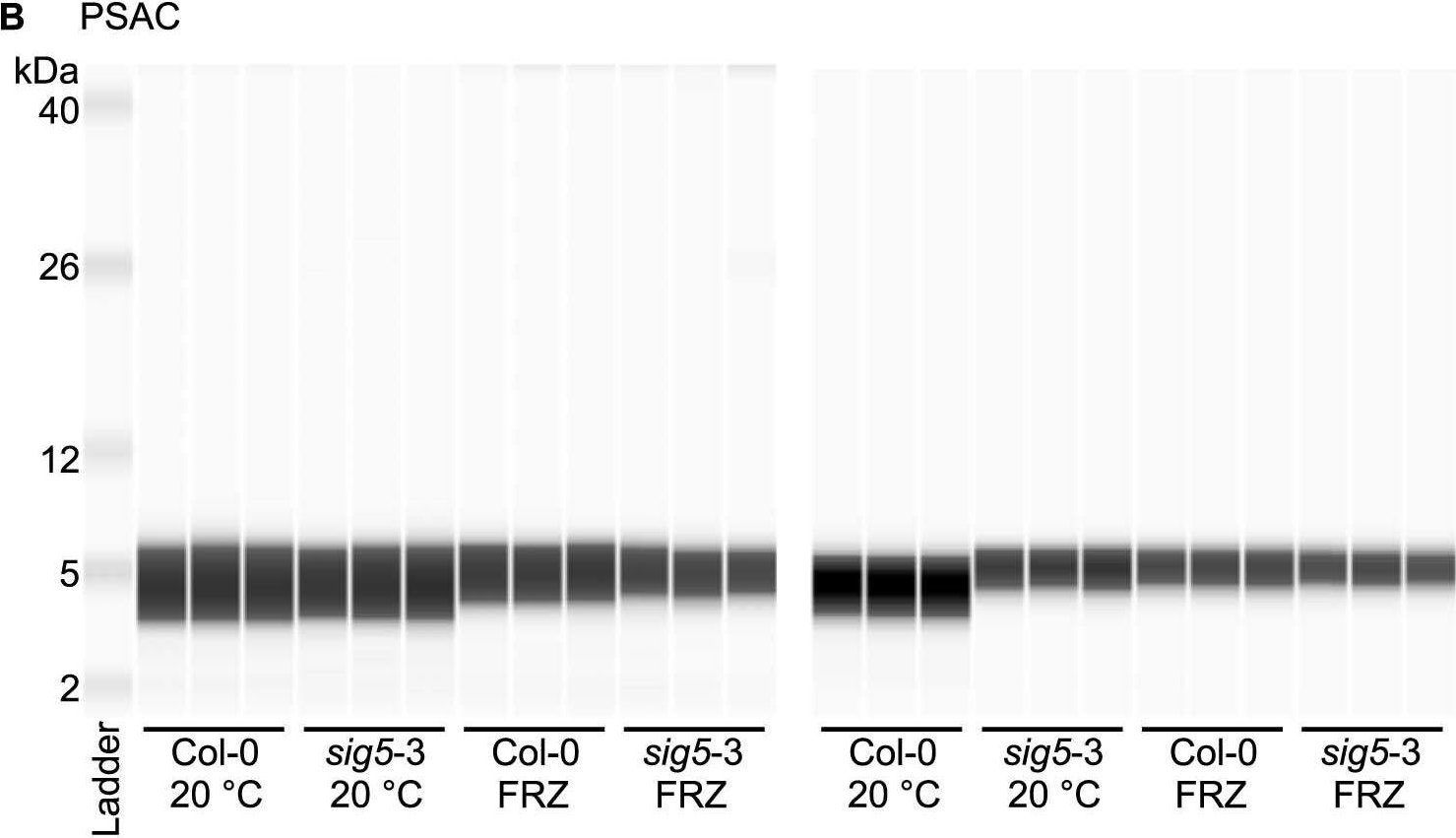
Reactant: Arabidopsis thaliana (Thale cress)
Application: Simple Western - Size
Pudmed ID: 36997687
Journal: Nat Plants
Figure Number: 5B
Published Date: 2023-04-01
First Author: Cano-Ramírez, D. L.
Impact Factor:
Open PublicationAltered photosystem protein abundance in sig5-3 mutant.Altered photosystem protein abundance in sig5-3 mutant. (A, B) Automated semi-quantitative immunoassay comparing (A) PSII D2 and (B) PSAC protein abundance between wild type (Col-0) and sig5-3 under control temperature conditions (20 °C) and after freezing at -8 °C for 6 h (FRZ). Analysis shows two independent experiments, each containing triplicate immunodetection analyses. (C) Coomassie blue-stained SDS-PAGE separation of 0.5 mg/mL total protein from leaf protein extracts, run as a single example to demonstrate consistent protein input into the automated semi-quantitative immunoassay when samples were prepared identically for immunodetection.
- Additional Information
-
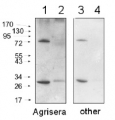
Additional information: Peptide target used to elicit this antibody is well conserved in all photoautotrophs except some cyanobacteria, some red algae and Cyanophora paradoxa, which contain a conserved substitution of a valine to an isoleucine. The performance of the antibodies has been confirmed against taxa containing both the valine and isoleucine variants.Example of a simulataneous western blot detection with RbcL, PsbA and PsaC antibodies.
More information about quantitative western blot using PsaC antibody can be found here.Additional information (application): In some species minor cross reactions with some larger proteins are seen. These may contain related iron-sulfur binding motifs. Therefore size verification of the reacting band is required. Due to the small size of the protein, care should be taken to differentiate between chemiluminescent signal from PsaC and non-specific signals from chlotophylls or lipids if pigment is retained near the bottom of the blot.For the most optimal results use:thylakoid membranes or PSI particles, solubilized in a SDS sample buffer (final concentrations: 63 mM Tris HCl, 10% glycerol, 2% SDS, 0.0025% bromophenol blue) with 2.5% beta-mercaptoethanol at 85C for 2 minutes. The samples were spun softly, then the supernatant loaded.
This product can be sold containing ProClin if requested. - Background
-
Background: PsaC is a conserved, chloroplast-encoded, Fe-S binding protein of approximately 10 kDa, present in all known Photosystem I complexes. It is located on the stromal side of the thylacoid membranes. PsaC coordinates the Fe–S clusters FA and FB through two cysteine-rich domains.
- Product Citations
-
Selected references: Xie et al, (2024). LpY3IP1 Enhances the drought and salt tolerance of perennial ryegrass by protecting the photosynthetic apparatus. Scientia Horticulturae Volume 338, 1 December 2024, 113645.Hani and Krieger-Liszkay (2024). Manganese deficiency alters photosynthetic electron transport in Marchantia polymorpha. Elsevier Plant Physiology and Biochemistry Available online 16 August 2024, 109042.
Frangedakis et al. (2024). MYB-related transcription factors control chloroplast biogenesis. Cell: DOI:https://doi.org/10.1016/j.cell.2024.06.039.
Wu et al (2023) Disruption of LEAF LESION MIMIC 4 affects ABA synthesis and ROS accumulation in rice
Cano-Ramirez et al. (2023) Low-temperature and circadian signals are integrated by the sigma factor SIG5. Nat Plants. 2023 Apr;9(4):661-672. doi: 10.1038/s41477-023-01377-1. Epub 2023 Mar 30.
Torrado, Connabeer, Rottig, et al. (2022) Directing cyanobacterial photosynthesis in a cytochrome c oxidase mutant using a heterologous electron sink. Plant Physiol. 2022;189(4):2554-2566. doi:10.1093/plphys/kiac204
Rredhi et al. (2023). The UV-A Receptor CRY-DASH1 Up- and Downregulates Proteins Involved in Different Plastidial Pathways. J Mol Biol. 2023 Sep 10:168271.doi: 10.1016/j.jmb.2023.168271.
Burlacot et al. (2022) Alternative photosynthesis pathways drive the algal CO2-concentrating mechanism. Nature 605, 366–371 (2022). https://doi.org/10.1038/s41586-022-04662-9
Ye et al. (2022) Effect of increased CO2 on iron-light-CO2 co-limitation of growth in a marine diatom, ASLO, Limnol. Oceanogr. 2022, 172-176
Rogowski et al. (2021) Light as a substrate: migration of LHCII antennas in extended Michaelis-Menten model for PSI kinetics. J Photochem Photobiol B. 2021 Dec;225:112336. doi: 10.1016/j.jphotobiol.2021.112336. Epub 2021 Oct 19. PMID: 34736069.
Levitan et al. (2019). Structural and functional analyses of photosystem II in the marine diatom Phaeodactylum tricornutum. Proc Natl Acad Sci U S A. 2019 Aug 27;116(35):17316-17322. doi: 10.1073/pnas.1906726116.
Zavrel et al. (2019). Quantitative insights into the cyanobacterial cell economy. Elife. 2019 Feb 4;8. pii: e42508. doi: 10.7554/eLife.42508.
Zhang et al. (2019). Proteomic responses to ocean acidification of the marine diazotroph Trichodesmium under iron-replete and iron-limited conditions. Photosynth Res. 2019 May 10. doi: 10.1007/s11120-019-00643-8.
Lupette et al. (2019). The architecture of lipid droplets in the diatom Phaeodactylum tricornutum. Algal Research Volume 38, March 2019, 101415.
Lima-Melo et al. (2019). Consequences of photosystem-I damage and repair on photosynthesis and carbon use in Arabidopsis thaliana. Plant J. 2018 Nov 29. doi: 10.1111/tpj.14177.
Steinbeck et al. (2018). Structure of a PSI-LHCI-cyt b6f supercomplex in Chlamydomonas reinhardtii promoting cyclic electron flow under anaerobic conditions. Proc Natl Acad Sci U S A. 2018 Oct 9;115(41):10517-10522. doi: 10.1073/pnas.1809973115.
Gonzaga Heredia-Martinez et al. (2018). Chloroplast damage induced by the inhibition of fatty acid synthesis triggers autophagy in Chlamydomonas. Plant Physiol, Sept. 2018.
Liu et al. (2018). Effects of PSII Manganese-Stabilizing Protein Succinylation on Photosynthesis in the Model Cyanobacterium Synechococcus sp. PCC 7002. Plant Cell Physiol. 2018 Jul 1;59(7):1466-1482. doi: 10.1093/pcp/pcy080.
Du et al. (2018). Galactoglycerolipid Lipase PGD1 Is Involved in Thylakoid Membrane Remodeling in Response to Adverse Environmental Conditions in Chlamydomonas. Plant Cell. 2018 Feb;30(2):447-465. doi: 10.1105/tpc.17.00446.
Cantrell and Peers (2017). A mutant of Chlamydomonas without LHCSR maintains high rates of photosynthesis, but has reduced cell division rates in sinusoidal light conditions. PLoS One. 2017 Jun 23;12(6):e0179395. doi: 10.1371/journal.pone.0179395.
Zang et al. (2017). Characterization of the sulfur-formation (suf) genes in Synechocystis sp. PCC 6803 under photoautotrophic and heterotrophic growth conditions. Planta. 2017 Jul 14. doi: 10.1007/s00425-017-2738-0.
Hu et al. (2017). The SUFBC2 D Complex is Required for the Biogenesis of All Major Classes of Plastid Fe-S Proteins. Plant J. 2017 Jan 19. doi: 10.1111/tpj.13483.
Yang-Er Chen et al. (2017). Responses of photosystem II and antioxidative systems to high light and high temperature co-stress in wheat. J. of Exp. Botany, Volume 135, March 2017, Pages 45–55.
Li et al. (2016). A Hard Day's Night: Diatoms Continue Recycling Photosystem II in the Dark. Front. Mar. Sci., 08 November 2016
Heinnickel et al. (2016). Tetratricopeptide repeat protein protects photosystem I from oxidative disruption during assembly. Proc Natl Acad Sci U S A. 2016 Mar 8;113(10):2774-9. doi: 10.1073/pnas.1524040113
Rozpadek et al. (2015). The fungal endophyte Epichloë typhina improves photosynthesis efficiency of its host orchard grass (Dactylis glomerata). Planta. 2015 Jun 10.
Subramanyam et al. (2014). Structural and functional changes of PSI-LHCI supercomplexes of Chlamydomonas reinhardtii cells grown under high salt conditions. Planta. 2010 Mar;231(4):913-22.
Dang et al. (2014). Combined Increases in Mitochondrial Cooperation and Oxygen Photoreduction Compensate for Deficiency in Cyclic Electron Flow in Chlamydomonas reinhardtii. Plant Cell. 2014 Jul 2. pii: tpc.114.126375. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
Oxygenic photosynthesis poster by prof. Govindjee and Dr. Shevela
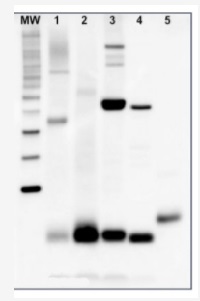
Z-scheme of photosynthetic electron transport by prof. Govindjee and Dr. Björn and Dr. ShevelaRubisco quantitation in plant and algal samples using Agrisera anti-PsaC global antibody and PsaC protein standard
Methodology: Plant samples are generally ground with liquid nitrogen in a mortar and pestle. The resulting powder is transferred to a plastic tube. Algal samples can be either concentrated by centrifugation or, preferably, by filtration onto glass fiber filters. Solubilization is performed in Agrisera protein extraction buffer (PEB, AS08 300) containing 0.1mg/mL PefaBloc SC (AEBSF) protease inhibitor (Roche). Disruption is most optimally obtained through flash freezing of the sample in liquid nitrogen alternated with thawing by sonication with a microtip. This process can be repeated depending on the toughness of the sample. The sample is adjusted to 50 mM dithiothreitol and heated to 70°C for 5 minutes. Samples are cooled and centrifuged briefly prior to electrophoresis.
Optimal quantitation is achieved using moderate sample loads per gel lane, generally 0.5 to 2.5 µg total protein, depending on the abundance of the target protein.
Electrophoresis and Immunoblotting: Once solubilized, the proteins can be separated electrophoretically in a number of systems. We obtain optimal results with the Invitrogen NuPAGE gel system using Bis-Tris 4-12% gradient gels. Proteins are separated in MES SDS running buffer according to the manufacturer’s recommendations at 200 V for 35 minutes. The gels are transferred to PVDF in the same apparatus, the SureLock XCell blot module, for 60 minutes at 30 V for a single gel or 80 minutes for a pair. Following transfer the blots are blocked in 2% blocking agent in Tris buffered saline with 0.1% Tween 20 (TBS-T) for 1 hour at room temperature with gentle agitation. The blot is incubated with primary antibody, usually at 1:25 000 to 1:50 000 diluted in 2% blocking agent, for 1 hour at room temperature. For quantitation a relatively high primary antibody:target protein ratio gives more reliable results than immunoblots at low ratios of primary antibody:target protein. The blot is washed extensively in TBS-T (twice briefly, once for 15 minutes and three times for five minutes). The blot is incubated with secondary antibody, for example goat anti-rabbit IgG horse radish peroxidase conjugated, at 1:50 000 in 2% blocking agent, for one hour at room temperature. The blot is washed as above and developed with chemiluminescence detection reagents as described by the manufacturer.
Quantitation: When quantitated standards are included on the blot, the samples can be quantitated using the available software. Excellent quantitation can be obtained with images captured on the Bio-Rad Fluor-S-Max or equivalent instrument using Bio-Rad QuantityOne software. The contour tool is used to select the area for quantitation and the values are background subtracted to give an adjusted volume in counts for each standard and sample. Using above protocol linear standard curves are generated over 1-1.5 orders of magnitude range in target load. It is important to note that immunodetections usually show a strongly sigmoidal signal to load response curve, with a region of trace detection of low loads, a pseudolinear range and a region of saturated response with high loads. For immunoquantitation it is critical that the target proteins in the samples and the standard curve fall within the pseudolinear range. Our total detection range using this protocol spans over 2 orders of magnitude, but the quantifiable range is narrower.
References:
MacKenzie et al (2005). Large reallocations of carbon, nitrogen and photosynthetic reductant among phycobilisomes, photosystems and Rubisco during light acclimation in Synechococcus elongatus are constrained in cells under low environmental inorganic carbon. Arch of Microbiol. 183: 190 - 202. Bouchard et al. (2006) UVB effects on the photosystem II-D1 protein of phytoplankton and natural phytoplankton communities. Photochem and Photobiol 82: 936-951. Morash et al. (2007) Macromolecular dynamics of the photosynthetic system over a seasonal developmental progression in Spartina alterniflora. Can J. of Bot. 85: 476-483(8)
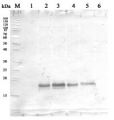
Application example

2 µg of total protein from (5-7) Prochlorococcus sp. extracted with Agrisera Protein Extration Buffer, PEB (AS08 300), (1-4) PsaC protein standard (AS02 042S) were separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Blots were blocked immediately following transfer in 2% blocking reagent in 20 mM Tris, 137 mM sodium chloride pH 7.6 with 0.1% (v/v) Tween-20 (TBS-T) for 1h at room temperature with agitation. Blots were incubated in the primary antibody at a dilution of 1: 50 000 for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated) diluted to 1:10 000 in 2% blocking solution for 1h at room temperature with agitation. The blots were washed as above and developed for 5 min with ECL detection reagent in mid picogram range, according the manufacturers instructions. Images of the blots were obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad). The native protein is significantly smaller than the tagged quantitation standard (lane 1-4). There is also a consistent cross-reaction with a larger FeS protein.
Recommended secondary antibodies: goat anti-rabbit HRP conjugated, goat anti-rabbit ALP conjugated
Recommended chemiluminescent detection reagent: AgriseraECLBright2 µg of total protein from (5-7) Prochlorococcus sp. extracted with Protein Extration Buffer, PEB (AS08 300), (1-4) PsaC protein standard (AS02 042S) were separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Blots were blocked immediately following transfer in 2% ECL Advance blocking reagent (GE Healthcare) in 20 mM Tris, 137 mM sodium chloride pH 7.6 with 0.1% (v/v) Tween-20 (TBS-T) for 1h at room temperature with agitation. Blots were incubated in the primary antibody at a dilution of 1: 50 000 for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated, from Abcam) diluted to 1:10 000 in 2% ECL Advance blocking solution for 1h at room temperature with agitation. The blots were washed as above and developed for 5 min with ECL Advance detection reagent according the manufacturers instructions. Images of the blots were obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad). The native protein is significantly smaller than the tagged quantitation standard (lane 1-4). There is also a consistent cross-reaction with a larger FeS protein. - Reviews:
-
Auroy Pascaline | 2021-06-07I used it on chlamydomonas. dilution 1/1000.
short migration and short transfer to have a better signalPiotr Rozpadek | 2014-03-24I confirm reactivity against Mesembryanthemum and Dactylis glomerata. crystallinum. Dillution: as in instruction