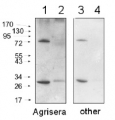
1
Anti-PsaD | PSI-D subunit of photosystem I
AS09 461 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Plants (monocots and dicots, conifers), moss: Physcomitrella patens, Chlamydomonas reinhardtii, Triticum aestivum, cyanobacteria
compartment marker of thylakoid membrane
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide 100% conserved in all known plant PsaD sequences including Arabidopsis thaliana PSI-D1 UniProt:Q9S7H1 , TAIR: At4g02770 and PSI-D2 UniProt: Q9SA56 , TAIR At1g03130 as well as Physcomitrella patens. The conservation in Chlamydomonas reinhardtii is high (14 of 16 aminoacids are identical).
Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Clear-native PAGE (CN-PAGE), Immunoprecipitation (IP), Western blot (WB) Recommended dilution: 1: 10 000 (CN-PAGE), 1 : 1000 - 1: 5 000 (WB) Expected | apparent MW: 17.9 | 20 (for Arabidopsis thaliana)
- Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Chlamydomonas reinhardtii, Dioxoniella giordanoi (red alga), Hordeum vulgare, Lactuca sativa, Nicotiana tabacum, Oryza sativa, Physcomitrium patens, Picea glauca, Pinus strobus, Oryza sativa, Physcomitrium patens, Spinacia oleracea, Synechocystis PCC 6803, Triticum aestivum, Triticale, Zea mays Predicted reactivity: Alge, Dicots, Catalpa bungei, Cucumis melo, Conifers, Cyanidioschyzon merolae, Bigelowiella natans, Nannochloropsis sp. , Phaeodactylum tricornutum, Phyla dulcis, Zosteria marina
Species of your interest not listed? Contact usNot reactive in: Synechococcus elongatus sp. PCC 7942
- Application Examples
-

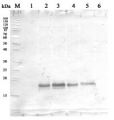
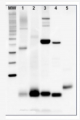
10 µg of total leaf protein extracted with PEB (AS08 300) from (1) Zea mays, (2) Chlamydomonas reinhardtii, and (3) Spinacia oleracea were separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 80 min (30V) to nitrocellulose. Filter was blocked 1h with 2% low-fat milk powder in TBS-T (0.1% TWEEN 20) and probed with anti-PsaD (AS09 461, 1:1000, 1h) and secondary anti-rabbit (1:40000, 1h) antibody (HRP conjugated) in TBS-T containing 2% low fat milk powder. Antibody incubations were followed by washings in TBS-T (15, +5, +5, +5 min). All steps were performed at RT with agitation. Signal was detected with chemiluminescent detection reagent using a GenoPlex Chemi CCD (accumulated signal 10 x 30s exposure, bin 2x2).
Total cellular (lanes 2 – 5) and membrane proteins (lanes 6 – 9) from various environmental isolated of Chlamydomonas reinhardtii were extracted with a buffer containing 62.5mM Tris-HCl pH 6.8, 10% glycerol, 2% SDS, 50mM DTT, 10mM NaF and 1% protease inhibitors (P9599, Sigma Aldrich) and denatured at 65°C for 5 min. Samples (0.25 µg of chlorophyll per lane) were separated on 12% SDS-PAGE containing 6M urea and blotted 1h to PVDF using tank transfer. Blots were blocked with 5% skim milk powder in TBS-T for 1h at room temperature (RT) with agitation. Blots were incubated in the primary antibody at a dilution of 1:5000 overnight at 4°C. The antibody solution was decanted and the blots were rinsed briefly once, then washed 3 times for 10 min in TBS-T at RT with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG HRP-conjugated, Agrisera AS09 602) diluted to 1:20 000 for 1h at RT with agitation. The blots were washed as above, developed for 5 min with chemiluminescent detection reagent and then imaged using a ChemiDoc MP imaging system and Image Lab software (Bio-Rad Laboratories). Exposure time was 10 seconds.
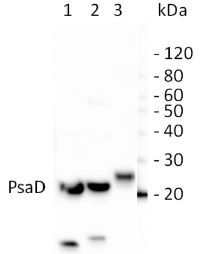
Courtesy of Kenneth Wilson, University of Saskatchewan, CanadaApplication examples: 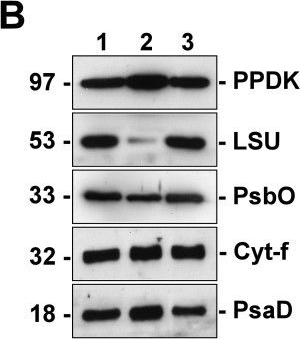
Reactant: Bienertia sinuspersici
Application: Western Blotting
Pudmed ID: 22394490
Journal: Plant Methods
Figure Number: 8B
Published Date: 2012-03-06
First Author: Lung, S. C., Yanagisawa, M., et al.
Impact Factor: 5.139
Open PublicationSilver staining and Western blot analyses of protein extracts from the isolated dimorphic chloroplasts. Two micrograms of proteins from total chloroplasts (lane 1), peripheral chloroplasts (lane 2) and central chloroplasts (lane 3) were separated on SDS/12% PAGE. Molecular weights are shown on the left in kilodaltons. (A) Total proteins were visualized by silver staining. Polypeptides of the highly abundant pyruvate orthophosphate dikinase (PPDK), Rubisco large-subunit (LSU) and small-subunit (SSU) are indicated. Other bands of significantly different intensities between the dimorphic chloroplasts are indicated by arrowheads. (B) Resolved proteins were electroblotted onto nitrocellulose membranes and probed with antibodies against PPDK, LSU, Photosystem II manganese-stabilizing protein (PsbO), cytochrome f (Cyt-f), or Photosystem I subunit II (PsaD). (C) The immunoblots were analyzed by densitometric quantification. The abundance of each protein in peripheral (P-Chls) and central (C-Chls) chloroplasts was calculated relative to that of total chloroplasts and shown with standard errors, after three (PsbO) or four (others) independent experiments.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 25835989
Journal: PLoS One
Figure Number: 7A
Published Date: 2015-04-04
First Author: Fristedt, R., Martins, N. F., et al.
Impact Factor: 2.942
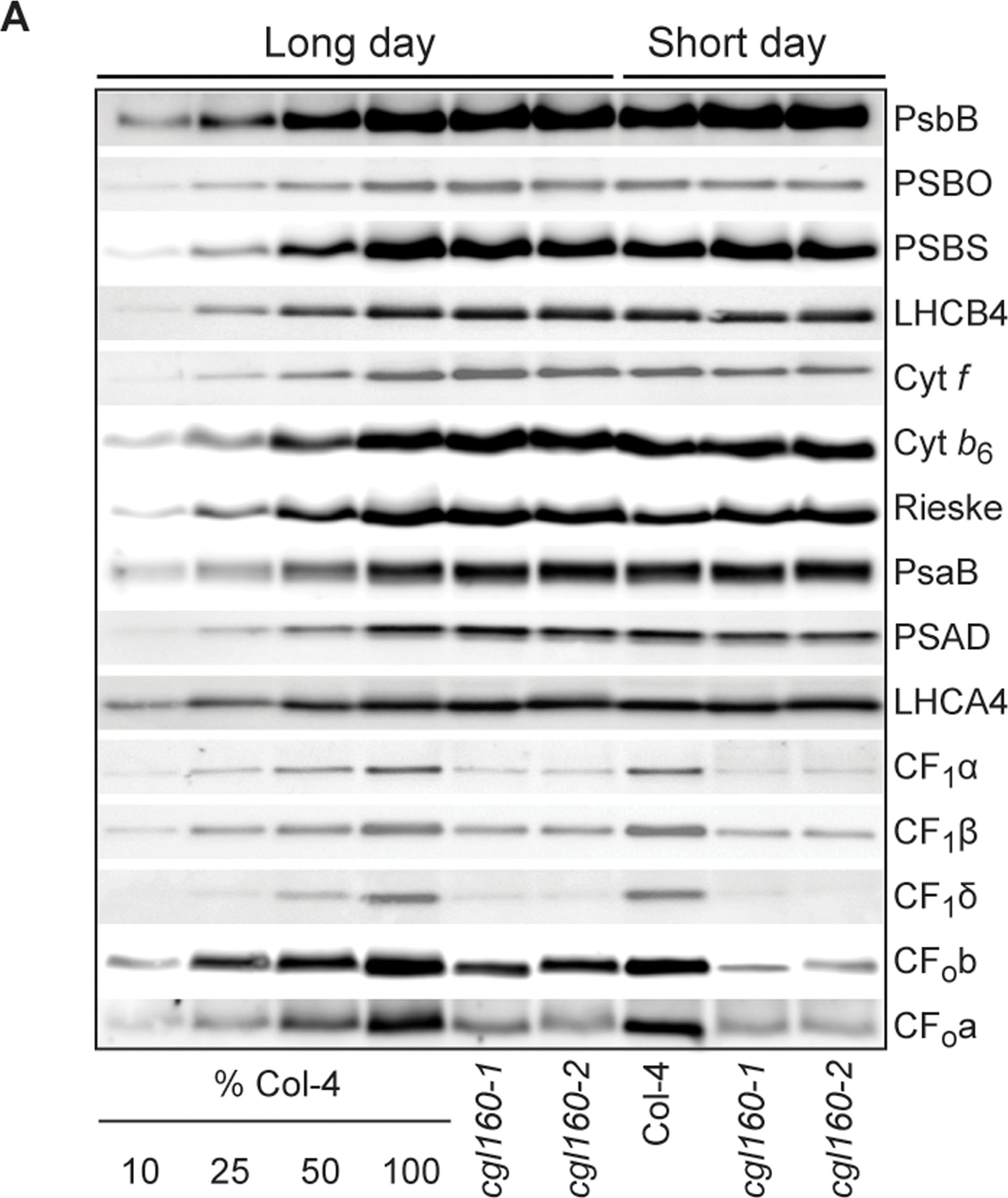
Open PublicationAltered protein accumulation and stability of the chloroplast ATP synthase in the cgl160 mutant visualized by immunoblotting.A. Immunoblots with antibodies against essential subunits of the photosynthetic protein complexes of wild-type (Col-4) Arabidopsis and the two cgl160 T-DNA insertion lines grown under long-day and short-day conditions. Isolated thylakoid membranes were used, and equal amounts of chlorophyll were loaded onto the SDS-PAGE gel. For approximate quantification, wild-type samples from long-day plants were diluted to 10%, 25% and 50%, respectively. Accumulation of PSII was probed with antibodies against PsbB and PSBO. Additionally, the PSBS protein involved in NPQ and the minor PSII antenna protein LHCB4 were probed. Accumulation of the cytochrome b6f complex was probed with antibodies against the essential subunits PetA (cytochrome f), PetB (cytochrome b6), and PETC (Rieske protein). Accumulation of PSI was probed with antibodies against the reaction center subunit PsaB and the stromal ridge subunit PsaD. ATP synthase accumulation was probed with antibodies against the CF1 subunits AtpA (CF1?), AtpB (CF1?) and AtpD (CF1?) and antibodies against the CF0 subunits AtpF (CF0b) and AtpI (CF0a). B. Loading difference estimation for immunoblotting CF1 between wild type and cgl160-1. To obtain similar immunoblotting signal three times more (15 ?g protein) was needed for cgl160-1 compared to wild type (5 ?g protein). C. Maintenance of CF1 was measured by incubating leaves from wild type and cgl160-1 in solution containing the plastid protein synthesis inhibitor chloramphenicol for the indicated time points. Protein extract was isolated and separated by SDS-PAGE, immunoblotted and probed with specific antibodies against CF1 and LHCB2.1. Three times more protein was loaded from the mutant to obtain equal level of CF1 immunoblotting signal, as specified in B.
Reactant: Nicotiana tabacum (Common tobacco)
Application: Western Blotting
Pudmed ID: 28180288
Journal: J Exp Bot
Figure Number: 5A
Published Date: 2017-02-01
First Author: Schöttler, M. A., Thiele, W., et al.
Impact Factor: 6.088
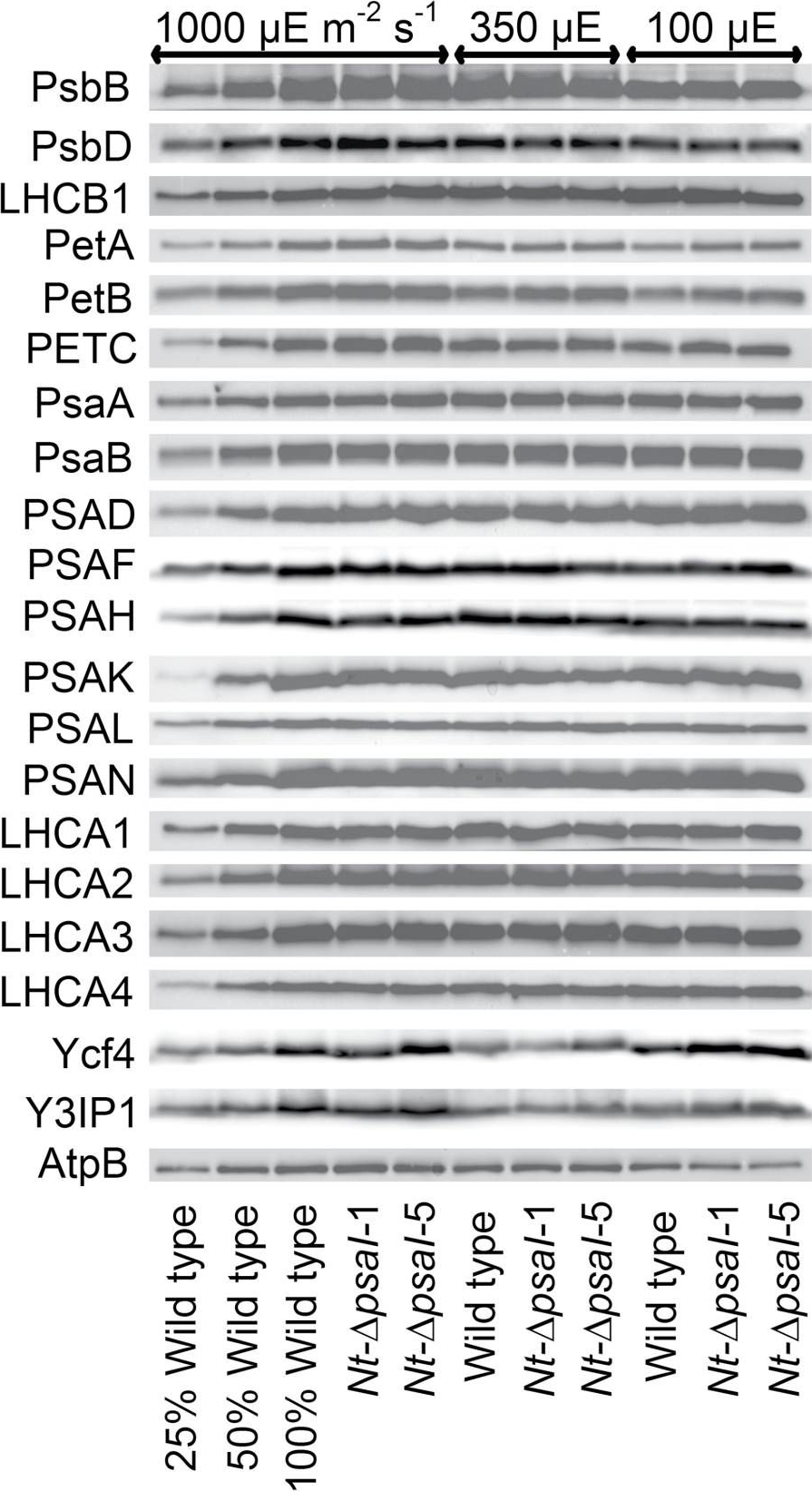
Open PublicationImmunoblot analysis of photosynthetic complex accumulation in wild-type tobacco and the two ?psaI lines grown under low, intermediate, and high-light conditions. Because the accumulation of most tested proteins was highest under high-light conditions, lanes one to three contain samples diluted to 25%, 50%, and a 100% sample of wild-type tobacco grown under high-light conditions, to allow for semi-quantitative determination of changes in protein abundance. Lanes four and five contain the two transplastomic lines grown at 1000 µE m?2 s?1. Lanes six to eight contain wild-type tobacco and the mutants grown at intermediate light intensities, and lanes nine to eleven contain samples grown at low light intensities. For PSII, the accumulation of the essential subunits PsbB (CP43) and PsbD (D2) and the LHCB1 antenna protein were determined, while for the cytochrome b6f complex, the accumulation of the essential redox-active subunits PetA (cytochrome f), PetB (cytochrome b6), and PETC (Rieske FeS protein) was tested. AtpB was probed as an essential subunit of the chloroplast ATP. For PSI, in addition to the three essential plastome-encoded subunits PsaA, PsaB, and PsaC, the accumulation of the nuclear-encoded subunits PSAD, PSAH, PSAK, PSAL, and PSAN and of the four LHCI proteins (LHCA1, LHCA2, LHCA3, LHCA4) was determined. Finally, we examined the accumulation of Ycf4, the chloroplast-encoded PSI-biogenesis factor encoded in the same operon as PsaI, and the nuclear-encoded assembly factor Y3IP1.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 28791032
Journal: Front Plant Sci
Figure Number: 2A
Published Date: 2017-08-10
First Author: Kohzuma, K., Froehlich, J. E., et al.
Impact Factor: 5.435
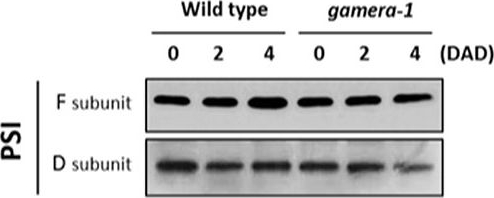
Open PublicationChanges in the protein levels of photosynthetic components under extended dark exposure in wild-type and gamera-1. Immunoblot detection of photosynthetic proteins from leaves of Ws and gamera-1 plants incubated after dark adaptation for 0, 2, and 4 days was examined. Specifically, essentially thylakoid fractions were assayed to determine the content of the following proteins: ?-subunit of ATP synthase; the D1 protein, OEC17, OEC23, and OEC33 of PSII; Cyt f and Rieske protein of the cytochrome b6f complex; and the F and D subunits of PSI, after extended dark treatment. Proteins were resolved via SDS-PAGE gel based on equal microgram chlorophyll per lane loading and processed as described in Section “Materials and Methods”. The Large subunit of RuBisco and LHCII stained with either CBB or Ponceau red, respectively, are presented here as loading controls. DAD indicates days after dark adaptation.
Reactant: Chlamydomonas reinhardtii (Green Alga)
Application: Western Blotting
Pudmed ID: 29875782
Journal: Front Plant Sci
Figure Number: 5E
Published Date: 2018-06-08
First Author: Zhang, W., Zhong, H., et al.
Impact Factor: 5.435
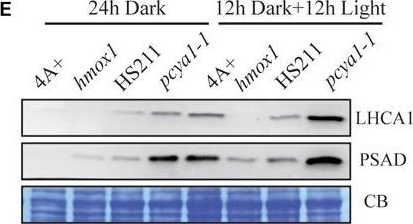
Open PublicationCharacterization of the CTE-disrupted pcya1-1 mutant. (A) Schematic diagram of an AphVIII insertion cassette into the PCYA1 locus in the pcya1-1 mutant. Black solid line represents 5?UTR and 3?UTR; black dotted line symbolizes intron; black box stands for exon and black arrows indicate the targeting sites of PCR primers. Purple, red, blue, and orange solid lines represent TP, NTE, FDBR and CTE regions of PCYA1, respectively. (B) PCR confirmation of the AphVIII insertion in pcya1-1. Genomic DNA was extracted from 4A+, HS211 and pcya1-1, respectively. PCR were performed using primers depicted in (A). HS211, the parental strain of pcya1-1. (C) Detection of CrPCYA1 expression in the dark or under light (?160 ?mol photons m-2s-1). Upper, immunoblot analysis of CrPCYA1 protein accumulation using antibody against CrPCYA1 mature protein; lower, Coomassie blue stain as an equal loading control. (D) Comparison of phototrophic growth phenotypes of pcya1-1 with 4A+, hmox1 and HS211 under constant low light (?60 ?mol photons m-2s-1), high light (?700 ?mol photons m-2s-1) and 12 h dark/12 h light cycle. (E) Accumulation of the photosystem I related proteins in pcya1-1, 4A+, HS211 and hmox1 in the dark or under light (?160 ?mol photons m-2s-1). Immunoblot analyses were performed with LHCA1 and PSAD antibodies. Coomassie blue stain (CB) as an equal loading control. Approximately 30 ?g total proteins were loaded in each lane.
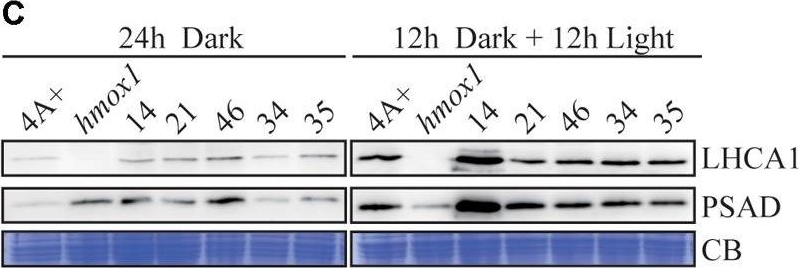
Reactant: Chlamydomonas reinhardtii (Green Alga)
Application: Western Blotting
Pudmed ID: 29875782
Journal: Front Plant Sci
Figure Number: 6C
Published Date: 2018-06-08
First Author: Zhang, W., Zhong, H., et al.
Impact Factor: 5.435
Open PublicationPCYA1 knockdown by artificial microRNA. (A) Analysis of CrPCYA1 protein accumulation in artificial microRNA-mediated PCYA1 knockdown lines. (B) Phototrophic growth of PCYA1 knockdown mutants were compared with 4A+ and hmox1 under constant low light, high light and dark/light cycle. Low light, ?60 ?mol photons m-2s-1; High light, ?700 ?mol photons m-2s-1. (C) Accumulation of representative photosystem I related proteins in PCYA1 knockdown mutants, 4A+ and hmox1 in the dark or under light (?160 ?mol photons m-2s-1). LHCA1 and PSAD antibodies were used for immunoblotting analyses. Around 30 ?g total proteins were loaded in each lane.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 32192026
Journal: Plants (Basel)
Figure Number: 1A
Published Date: 2020-03-17
First Author: Teubner, M., Lenzen, B., et al.
Impact Factor: None
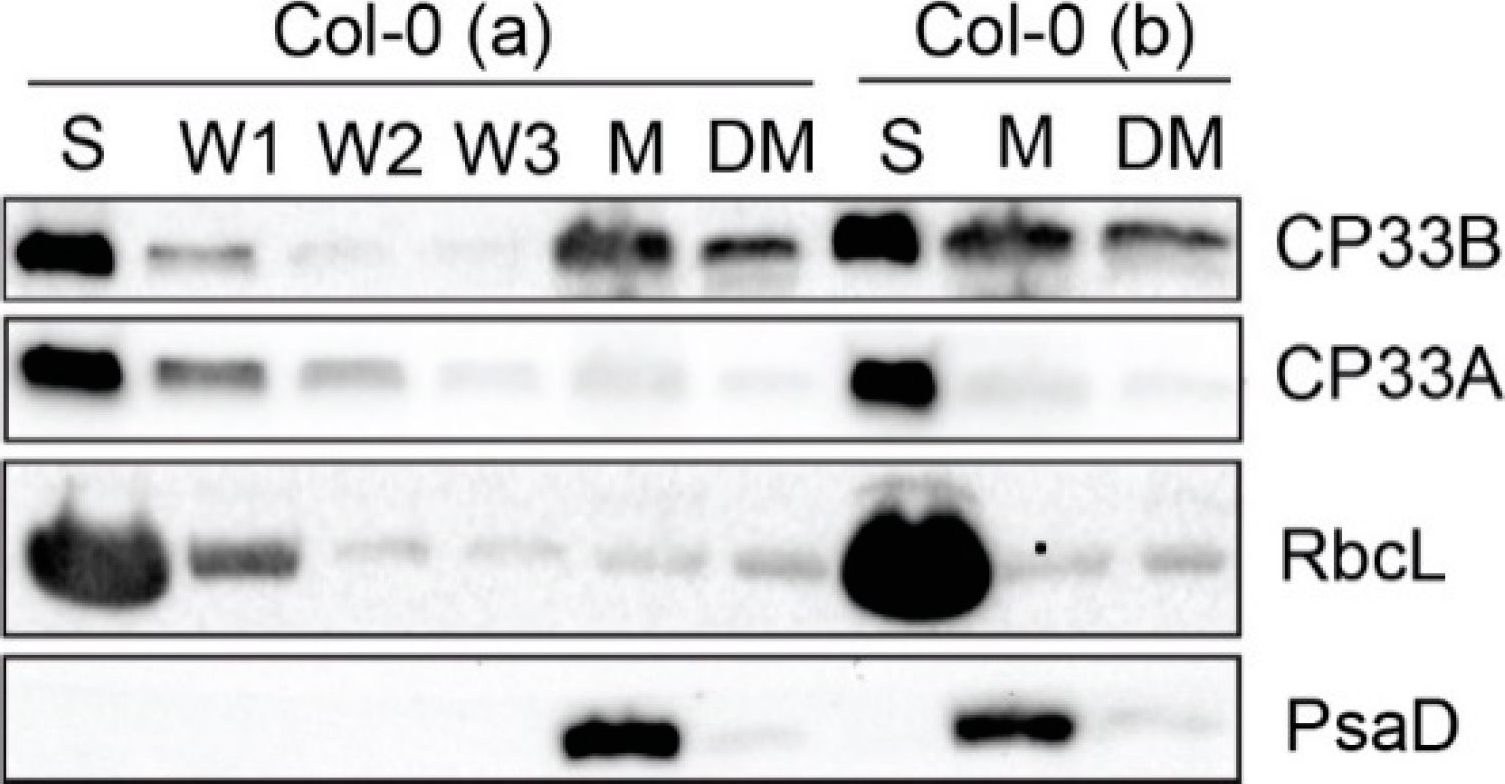
Open PublicationCP33B localizes to stroma and membrane fraction of chloroplasts. Chloroplasts of two wild type replicates (Col-0 a and b) were separated into stroma (S) and membranes (M). Membranes were washed 5 times. A membrane aliquot was treated with 0.5% sodium deoxycholate and the supernatant was harvested after treatment (DM). From each fraction, including the first three washing steps (W1, W2, W3) of Col-0 (a), equal volume fractions were separated electrophoretically by SDS PAGE and transferred to a nitrocellulose membrane. Besides the immunological detection of CP33A (antibody peptide a) and CP33B, antibodies for RbcL (large subunit of RuBisCO (ribulose 1.5 bisphosphate carboxylase/oxygenase)) and PsaD (subunit of photosystem I) served as stroma and membrane markers, respectively. The controls (RbcL, CP33A, PsaD) are reprinted with permission from Wiley; original Figure 2c in [8]; © 2020 John Wiley & Sons Ltd, Hoboken, New Jersey, USA.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 32269582
Journal: Front Plant Sci
Figure Number: 7B
Published Date: 2020-04-10
First Author: Pralon, T., Collombat, J., et al.
Impact Factor: 5.435
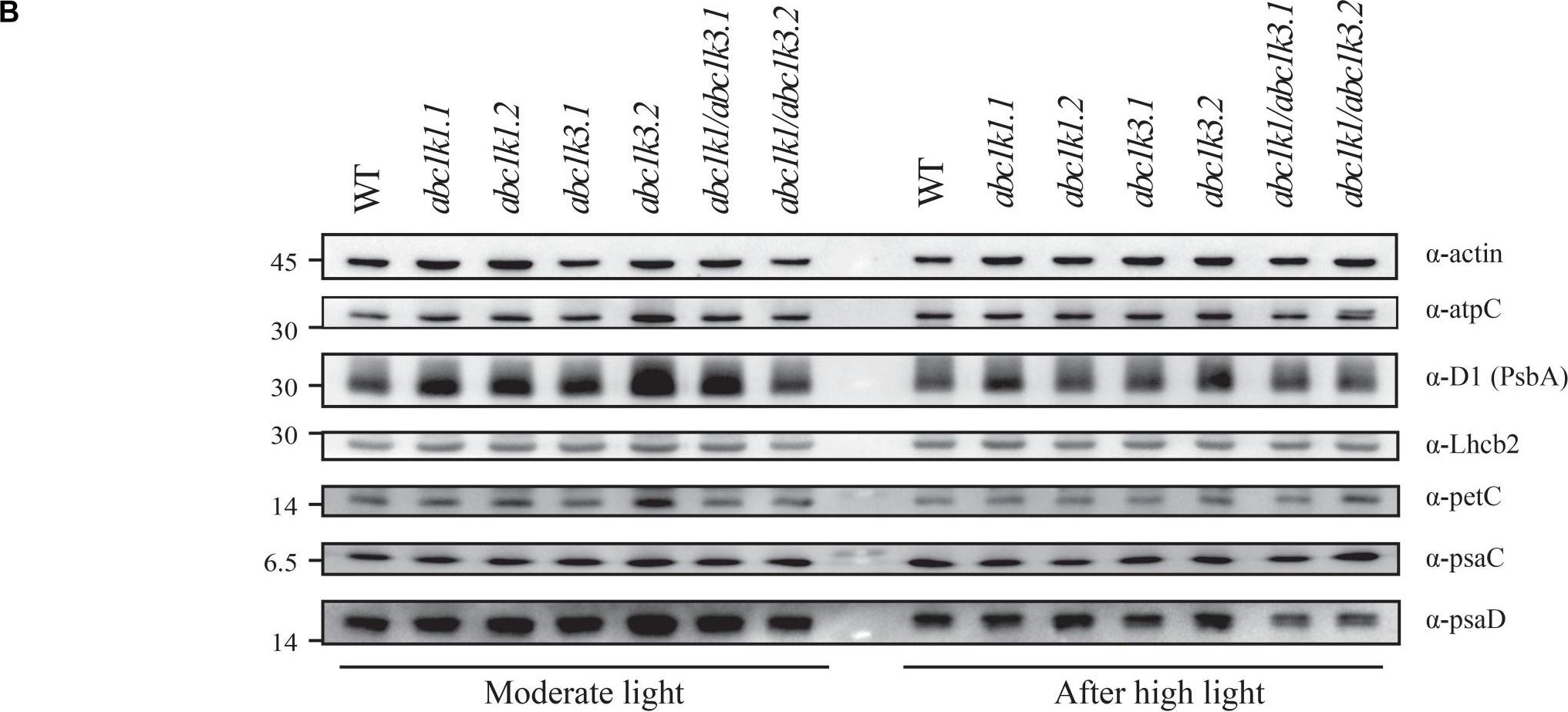
Open PublicationDouble mutant maintains thylakoid protein phosphorylation and state transitions after high light. (A) Total protein extracts of wild type (WT), abc1k1.1, -2, abc1k3.1, -2, and abc1k1/abc1k3.1, -2 light-exposed leaves were separated by SDS PAGE, transferred on nitrocellulose membrane and decorated with anti-phosphothreonine antibody. The main thylakoid phospho-proteins are indicated on the right according to their size. Core photosystem II proteins D1 (PsbA) and D2 (PsbD) are indicated together due their poor resolution. (B) The accumulation of the principal photosynthetic complexes was assessed using antibodies against specific subunits of each complex: anti-Lhcb2 for the major LHCII, anti-D1 (PsbA) for PSII, anti-PetC for cytochrome b6f, anti-PsaD and anti-PsaC for PSI, and anti-AtpC for ATP synthase. Actin signal is shown as a loading control. (C) Fluorescence quenching related to the state transitions (qT) of wild type (WT), abc1k1.1, -2, abc1k3.1, -2, and abc1k1/abc1k3.1, -2 under moderate light (120 ?mol of photons m–2 s–1) (ML) and after 3 h of high light (500 ?mol of photons m–2 s–1) (HL). qT was calculated from the maximal chlorophyll fluorescence measured after 10 min exposure to red light (660 nm) supplemented with far-red illumination (720 nm) “State 1” (FMST1) or to pure red light “State 2” (FMST2). Quenching related to state transition was calculated as qT = (FMST1 – FMST2)/FM. Each value represents the average of a pot containing 2–3 plants. Superscript letters are used to indicate statistically different groups (p < 0.05) by paired Student’s t-test.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 33629953
Journal: Elife
Figure Number: 6A
Published Date: 2021-02-25
First Author: Pipitone, R., Eicke, S., et al.
Impact Factor: 7.448
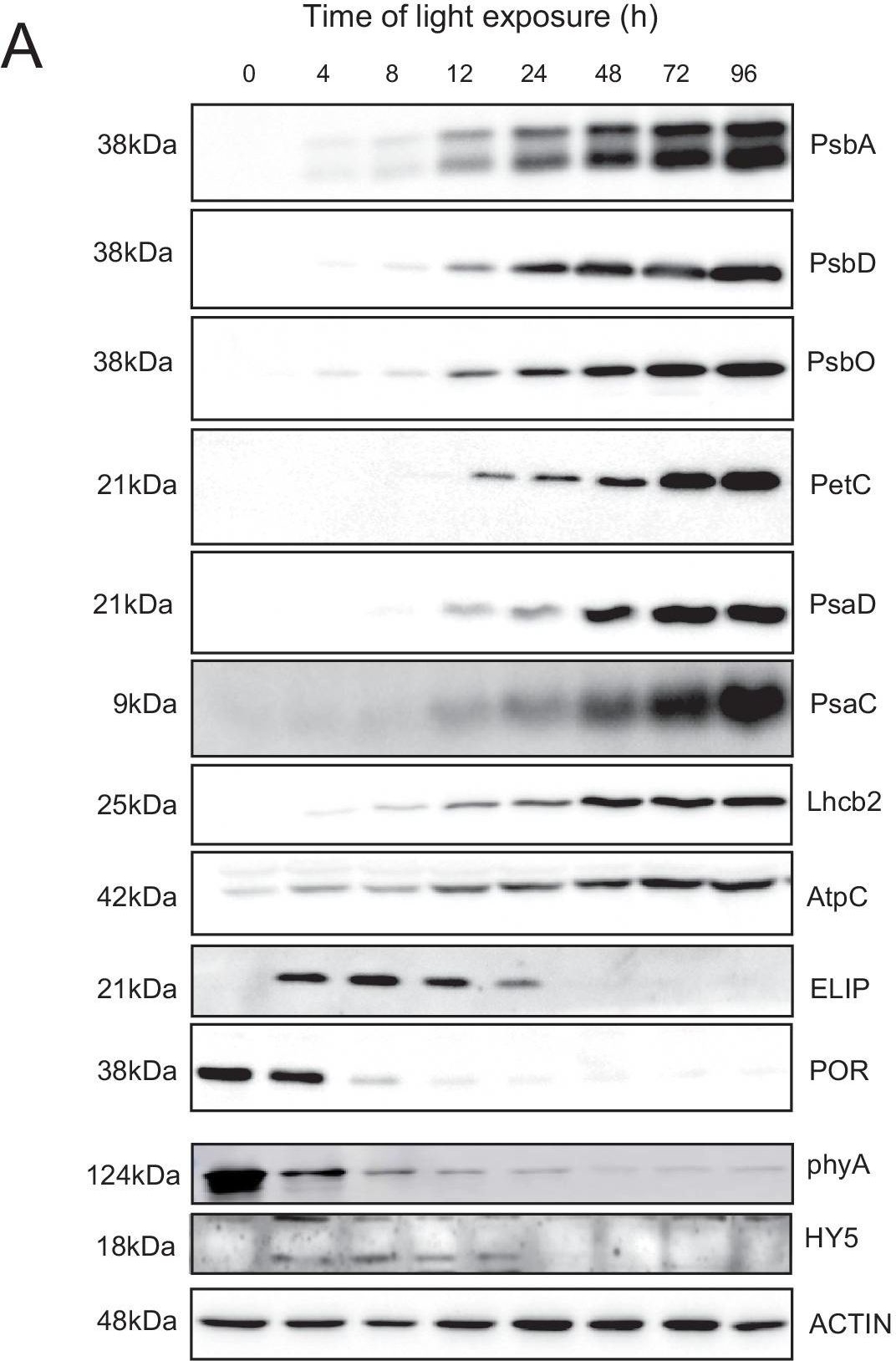
Open PublicationAccumulation dynamics of photosynthesis-related proteins during de-etiolation.Three-day-old etiolated seedlings of Arabidopsis thaliana were illuminated for 0 hr (T0), 4 hr (T4), 8 hr (T8), 12 hr (T12), 24 hr (T24), 48 hr (T48), 72 hr (T72), and 96 hr (T96) under white light (40 µmol/m2/s). (A) Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membrane and immunodetected with antibodies against PsbA, PsbD, PsbO, PetC, PsaD, PsaC, Lhcb2, AtpC, ELIP, POR, phyA, HY5, and ACTIN proteins. (B–C) Quantification of PsbA, PetC, and PsaC during de-etiolation. Heatmap (B) was generated after normalization of the amount of each protein relative to the last time point (T96). Graph (C) corresponds to the absolute quantification of proteins at T96. Error bars indicate ± SD (n = 3). Quantification of photosystem-related proteins during de-etiolation is detailed in Figure 6—figure supplement 1.Figure 6—source data 1.Quantitative data for immunoblot analysis.Quantitative data for immunoblot analysis.Quantification of photosynthesis-related proteins.(A) Immunodetection of PsbA, PetC, and PsaC during de-etiolation. Dilutions were used for the later time points to avoid saturation of the signal. (B) Different bands were detected by Amersham Imager program and quantified by Image QuantTL (Amersham). (C) Calibration curves were created using recombinant proteins (Agrisera). Calibration curve composition: PsbA 10 ng (A; lane a), 5 ng (b), 2.5 ng (c), and 1.25 ng (d); PetC 10 ng (e), 5 ng (f), 2.5 ng (g), and 1.25 ng (h); PsaC 3 ng (i), 1.5 ng (l), 0.75 ng (m), and 0.325 ng (n). Data indicate mean ± SD (n = 3–4). Raw data and calculations are shown in Figure 6—source data 1.
- Additional Information
-
Additional information: PsaD has frequently been used as a marker for intact PSI reaction centers.
This product can be sold containing proclin if requested.Additional information (application): This antibody is a replacement for former product, anti-PsaD AS04 046
Contains 0.1% ProClin. - Background
-
Background: PsaD (PSI-D) is a core subunit of photosystem I highly conserved in all photosynthetic organisms (including bacteria with Fe-S type reaction centers). In eukaryots its encoded by 1 to 2 nuclear gene(s) and imported as a precursor into the chloroplast. In the thylakoid membrane it associates with PsaA and PsaB on the stromal site of the PSI core forming the Fd-docking site. PsaD is also required for the stable assembly of PsaC.
- Product Citations
-
Selected references: Collombat et al. (2025). Arabidopsis conditional photosynthesis mutants abc1k1 and var2 accumulate partially processed thylakoid preproteins and are defective in chloroplast biogenesis. Commun Biol . 2025 Jan 22;8(1):111. doi: 10.1038/s42003-025-07497-y.
Hang et al. (2024). HOT3/eIF5B1 confers Kozak motif-dependent translational control of photosynthesis-associated nuclear genes for chloroplast biogenesis. Nat Commun. 2024 Nov 14;15(1):9878. doi: 10.1038/s41467-024-54194-1.
Penzler et al. (2024). A pgr5 suppressor screen uncovers two distinct suppression mechanisms and links cytochrome b6f complex stability to PGR5. Plant Cell. 2024 Mar 27:koae098. doi: 10.1093/plcell/koae098.
Völkner et al. (2024). Evidence for partial functional overlap of KEA and MSL transport proteins in the chloroplast inner envelope of Arabidopsis thaliana. FEBS Lett. 2024 Aug;598(15):1877-1887. doi: 10.1002/1873-3468.14887.
Uflewski et al. (2024). The thylakoid proton antiporter KEA3 regulates photosynthesis in response to the chloroplast energy status. Nat Commun. 2024 Mar 30;15(1):2792. doi: 10.1038/s41467-024-47151-5.
Okegawa et al. (2023). x- and y-type thioredoxins maintain redox homeostasis on photosystem I acceptor side under fluctuating light. Plant Physiol. 2023 Nov 22;193(4):2498-2512.doi: 10.1093/plphys/kiad466.
Hao and Malnoë (2023). A Simple Sonication Method to Isolate the Chloroplast Lumen in Arabidopsis thaliana.Bio Protoc. 2023 Aug 5; 13(15): e4756.
Jin et al. (2023) Dual roles for CND1 in maintenance of nuclear and chloroplast genome stability in plants. Cell Rep. 2023 Mar 28;42(3):112268. doi: 10.1016/j.celrep.2023.112268. Epub 2023 Mar 17.
Maeda et al. (2022). Characterization of photosystem II assembly complexes containing ONE-HELIX PROTEIN1 in Arabidopsis thaliana. J Plant Res. 2022 Mar;135(2):361-376. doi: 10.1007/s10265-022-01376-x. Epub 2022 Feb 10. PMID: 35146632.(CN-PAGE)
Gao Y et al. (2022) Chloroplast translational regulation uncovers nonessential photosynthesis genes as key players in plant cold acclimation. Plant Cell. 2022 Apr 26;34(5):2056-2079. doi: 10.1093/plcell/koac056. PMID: 35171295; PMCID: PMC9048916.
Espinoza-Corral & Lundquist. (2022) The plastoglobule-localized protein AtABC1K6 is a Mn2+-dependent kinase necessary for timely transition to reproductive growth. J Biol Chem. 2022 Apr;298(4):101762. doi: 10.1016/j.jbc.2022.101762. Epub 2022 Feb 22. PMID: 35202657; PMCID: PMC8956952.
Xiong et al. (2022) a chloroplast nucleoid protein of bacterial origin linking chloroplast transcriptional and translational machineries, is required for proper chloroplast gene expression in Arabidopsis thaliana. Nucleic Acids Res. 2022 Jun 23;50(12):6715–34. doi: 10.1093/nar/gkac501. Epub ahead of print. PMID: 35736138; PMCID: PMC9262611.
Ivanov et al. (2022) The decreased PG content of pgp1 inhibits PSI photochemistry and limits reaction center and light-harvesting polypeptide accumulation in response to cold acclimation. Planta 255, 36 (2022). https://doi.org/10.1007/s00425-022-03819-0
Fukura et al. (2021) Enrichment of chlorophyll catabolic enzymes in grana margins and their cooperation in catabolic reactions. J Plant Physiol. 2021 Nov;266:153535. doi: 10.1016/j.jplph.2021.153535. Epub 2021 Sep 25. PMID: 34607178.
Fattore et al. (2021). Acclimation of photosynthetic apparatus in the mesophilic red alga Dixoniella giordanoi. Physiol Plant. 2021 Nov;173(3):805-817. doi: 10.1111/ppl.13489. Epub 2021 Jul 5. PMID: 34171145; PMCID: PMC8596783.
Chen et al. (2021)Degradation of the photosystem II core complex is independent of chlorophyll degradation mediated by Stay-Green Mg2+ dechelatase in Arabidopsis,Plant Science,Volume 307,2021,110902,ISSN 0168-9452,https://doi.org/10.1016/j.plantsci.2021.110902.
Pipitone et al. (2021). A multifaceted analysis reveals two distinct phases of chloroplast biogenesis during de-etiolation in Arabidopsis. Elife. 2021 Feb 25;10:e62709. doi: 10.7554/eLife.62709. PMID: 33629953; PMCID: PMC7906606.
Kamea et al. (2021). Substitution of deoxycholate with the amphiphilic polymer amphipol A8-35 improves the stability of large protein complexes during native electrophoresis. Plant Cell Physiol. 2021 Jan 5:pcaa165. doi: 10.1093/pcp/pcaa165. Epub ahead of print. PMID: 33399873.
Aso et al. (2021). Unique peripheral antennas in the photosystems of the streptophyte alga Mesostigma viride. Plant Cell Physiol. 2021 Jan 8:pcaa172. doi: 10.1093/pcp/pcaa172. Epub ahead of print. PMID: 33416834.
Tang el al. (2020). OsNSUN2-Mediated 5-Methylcytosine mRNA Modification Enhances Rice Adaptation to High Temperature. Dev Cell. 2020 May 4;53(3):272-286.e7. doi: 10.1016/j.devcel.2020.03.009.
Wang et al. (2020) Rerouting of ribosomal proteins into splicing in plant organelles. BioRxiv, DOI: 10.1101/2020.03.03.974766 .BN-PAGE
Teubner et al. (2020). The chloroplast ribonucleoprotein CP33B quantitatively binds the psbA mRNA. doi.org/10.1101/2020.02.11.944249
Storti et al. (2020). The activity of chloroplast NADH dehydrogenase-like complex influences the photosynthetic activity of the moss Physcomitrella patens. doi.org/10.1101/2020.01.29.924597
Xu et al. (2019). VENOSA4, a Human dNTPase SAMHD1 Homolog, Contributes to Chloroplast Development and Abiotic Stress Tolerance.
Chen et al. (2019). Effects of Stripe Rust Infection on the Levels of Redox Balance and Photosynthetic Capacities in Wheat. Int J Mol Sci. 2019 Dec 31;21(1). pii: E268. doi: 10.3390/ijms21010268.
Furukawa et al. (2019). Formation of a PSI–PSII megacomplex containing LHCSR and PsbS in the moss Physcomitrella patens. J Plant Res https://doi.org/10.1007/s10265-019-01138-2.
Storti et al. (2018). Role of cyclic and pseudo-cyclic electron transport in response to dynamic light changes in Physcomitrella patens. Plant Cell Environ. 2018 Nov 29. doi: 10.1111/pce.13493.
Mao et al. (2018). Comparison on Photosynthesis and Antioxidant Defense Systems in Wheat with Different Ploidy Levels and Octoploid Triticale. Int J Mol Sci. 2018 Oct 2;19(10). pii: E3006. doi: 10.3390/ijms19103006.
Nama et al. (2018). Non-photochemical quenching-dependent acclimation and thylakoid organization of Chlamydomonas reinhardtii to high light stress. Photosynth Res. 2018 Jul 7. doi: 10.1007/s11120-018-0551-7.
Gao et al. (2018). Effect of green light on the amount and activity of NDH-1–PSI supercomplex in Synechocystis sp. strain PCC 6803. Photosynthetica (2018) 56: 316. https://doi.org/10.1007/s11099-018-0790-z.
Kong et al. (2018) Interorganelle Communication: Peroxisomal MALATE DEHYDROGENASE2 Connects Lipid Catabolism to Photosynthesis through Redox Coupling in Chlamydomonas. Plant Cell. 2018 Aug;30(8):1824-1847. doi: 10.1105/tpc.18.00361
Li et al. (2018). Modulating plant growth-metabolism coordination for sustainable agriculture. Nature. 2018 Aug 15. doi: 10.1038/s41586-018-0415-5.
Du et al. (2018). Galactoglycerolipid Lipase PGD1 Is Involved in Thylakoid Membrane Remodeling in Response to Adverse Environmental Conditions in Chlamydomonas. Plant Cell. 2018 Feb;30(2):447-465. doi: 10.1105/tpc.17.00446.
Schottler et al. (2017). The plastid-encoded PsaI subunit stabilizes photosystem I during leaf senescence in tobacco. J Exp Bot. 2017 Feb 1;68(5):1137-1155. doi: 10.1093/jxb/erx009.
Merry et al. (2017). A comparison of pine and spruce in recovery from winter stress; changes in recovery kinetics, and the abundance and phosphorylation status of photosynthetic proteins during winter. Tree Physiol. 2017 Sep 1;37(9):1239-1250. doi: 10.1093/treephys/tpx065.
Ge at al. (2017). Translating Divergent Environmental Stresses into a Common Proteome Response through the Histidine Kinase 33 (Hik33) in a Model Cyanobacterium. Mol Cell Proteomics. 2017 Jul;16(7):1258-1274. doi: 10.1074/mcp.M116.068080.
Yang-Er Chen et al. (2017). Responses of photosystem II and antioxidative systems to high light and high temperature co-stress in wheat. J. of Exp. Botany, Volume 135, March 2017, Pages 45–55.
Gerotto et al. (2016). Flavodiiron proteins act as safety valve for electrons in Physcomitrella patens. PNAS DOI 10.1073.
Heinnickel et al. (2016). Tetratricopeptide repeat protein protects photosystem I from oxidative disruption during assembly. Proc Natl Acad Sci U S A. 2016 Mar 8;113(10):2774-9. doi: 10.1073/pnas.1524040113
Fristedt et al. (2015). The thylakoid membrane protein CGL160 supports CF1CF0 ATP synthase accumulation in Arabidopsis thaliana. PLoS One. 2015 Apr 2;10(4):e0121658. doi: 10.1371/journal.pone.0121658.
Fujii et al. (2015). Photoprotection vs Photoinhibition of Photosystem II in Transplastomic Lettuce (Lactuca sativa) Dominantly Accumulating Astaxanthin. Plant Cell Physiol. 2015 Dec 7. pii: pcv187.
Daddy et al. (2015). A novel high light-inducible carotenoid-binding protein complex in the thylakoid membranes of Synechocystis PCC 6803. Sci Rep. 2015 Mar 30;5:9480. doi: 10.1038/srep09480.
Armbruster et al. (2014). Ion antiport accelerates photosynthetic acclimation in fluctuating light environments. Nat Commun. 2014 Nov 13;5:5439. doi: 10.1038/ncomms6439
Qin et al. (2014). Isolation and characterization of a PSI-LHCI super-complex and its sub-complexes from a siphonaceous marine green alga, Bryopsis Corticulans. Photosynth Res. 2014 Sep 12.
Cheng and He (2014). PfsR Is a Key Regulator of Iron Homeostasis in Synechocystis PCC 6803. PLoS One. 2014 Jul 10;9(7):e101743. doi: 10.1371/journal.pone.0101743. eCollection 2014.
Tomizioli et al. (2014). Deciphering thylakoid sub-compartments using a mass spectrometry-based approach. Mol Cell Proteomics. 2014 May 28. pii: mcp.M114.040923.
Lang et al. (2011).Simultaneous isolation of pure and intact chloroplasts and mitochondria from moss as the basis for sub-cellular proteomics. Plant Cell Rep. 2011 Feb;30(2):205-15.doi: 10.1007/s00299-010-0935-4. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
Oxygenic photosynthesis poster by prof. Govindjee and Dr. Shevela
Z-scheme of photosynthetic electron transport by prof. Govindjee and Dr. Björn and Dr. Shevela - Reviews:
-
Marie Bertrand | 2023-07-26We used this antibody on Chlamydomonas andget a verry strong intensity with 1:1000 dilution. Good product.Ben Field | 2020-02-19A good antibody that gives a strong band on Arabidopsis total proteins (15ug) at a dilution of 1:2000 in TBST 5% milk.Amy Verhoeven | 2020-02-07We used this antibody on both Eastern white pine (Pinus strobus) and White spruce (Picea glauca) with success in western blotting. We used 20 ug protein per lane and a dilution of 1:10,000. We developed using ECL.
Merry et al. 2017 Phys Plant 37:1239-1250 https://doi.org/10.1093/treephys/tpx065Julia Hamm | 2018-11-21Clear signal in Synechocystis with a dilution 1:1000| 2015-01-19We carried out a WB on thylakoid membrane proteins using a dilution of 1:1000 (ECL). It gave a very strong single band.
Accessories
AS10 939 | Clonality: Polyclonal | Host: Rabbit | Reactivity: [global antibody] for higher plants, algae, cyanobacteria, diatoms
Benefits of using this antibody
50% discount on matching standard/positive control
AS04 042S PsaC | PSI positive control/quantitation standard
Use promotional code: Stand50