1


Anti-PetC | Rieske iron-sulfur protein of Cyt b6/f complex
AS08 330 | Clonality: Polyclonal | Host: Rabbit | Reactivity: A. thaliana, B. rapa subsp. chinensis, C. reinhardtii, E. crus-galli, Euglena sp., H. pluvialis, N. tabacum, P. miliaceum, P. sativum, S. oleracea, Synechococcus PCC 7942, Synechocystis sp. PCC 6803, Thalassiosira guillardii, Z. mays
Benefits of using this antibody
50% discount on matching standard/positive control
AS08 330S PetC | Positive control/quantitation standard
Use promotional code: Stand50
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide which shows strong conservation across higher plants including Arabidopsis thaliana UniProt:Q9ZR03, TAIR:At4g03280, Chlamydomonas reinhardtii P49728 and Synechococcus sp. Q5N5B0 Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Blue Native PAGE (BN-PAGE), Western blot (WB) Recommended dilution: 1 : 5000-1 : 10 000 (BN-PAGE), (WB) Expected | apparent MW: 23 kDa
- Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Brassica rapa subsp. chinensis, Chlamydomonas reinhardtii, Echinola crus-galli, Euglena sp., Haematococcus pluvialis, Nicotiana tabacum, Panicum miliaceum, Pisum sativum, Setaria viridis, Spinacia oleracea, Synechococcus PCC 7942, Synechocystis sp. PCC 6803, Thalassiosira guillardii, Zea mays Predicted reactivity: Acetabularia acetabulum, Brachypodium distachyon, cyanobacteria, Calothrix sp. PCC 7507, Catalpa bungei, Cicer arietinum, Crocosphaera watsonii, Cynodon dactylon, Gossypium raimondii , Hordeum vulgare, Lyngbya aestuarii,Microcystis aeruginosa, Nannochloropsis gaditana, Nicotiana benthamiana, Pisum sativum, Ricinus communis , Saccharum hybrid cultivar ROC22, Selaginella moellendorffii, Solanum tuberosum, Sorghum bicolor, Oryza sativa, Physcomitrium patens, Phormidesmis priestleyi, Populus trichocarpa, Sonneratia alba, Triticum aestivum, Zostera marina, Vitis vinifera
Species of your interest not listed? Contact usNot reactive in: Candidia albicans - Application Examples
-
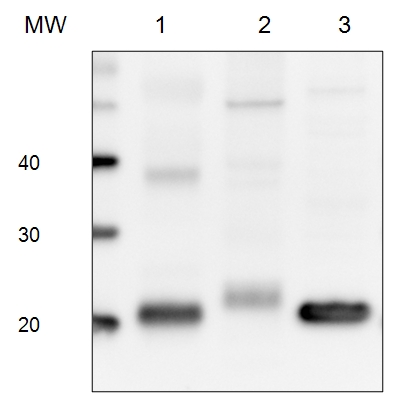
5 µg of total protein from (1) Arabidopsis thaliana leaf extracted with Protein Extration Buffer, PEB (AS08 300), (2) Euglena sp. extracted with PEB, (3) Synechococcus elongatus whole cell extracted with PEB, were separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Blots were blocked immediately following transfer in 2% blocking reagent 0.1% (v/v) Tween-20 (TBS-T) for 1h/RT with agitation. Blots were incubated in the primary antibody at a dilution of 1: 10 000 for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated) diluted to 1:50 000 for 1h/RT with agitation. The blots were washed as above and developed for 5 min withchemiluminescence detection reagent according the manufacturers instructions. Images of the blots were obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad).
Application examples: 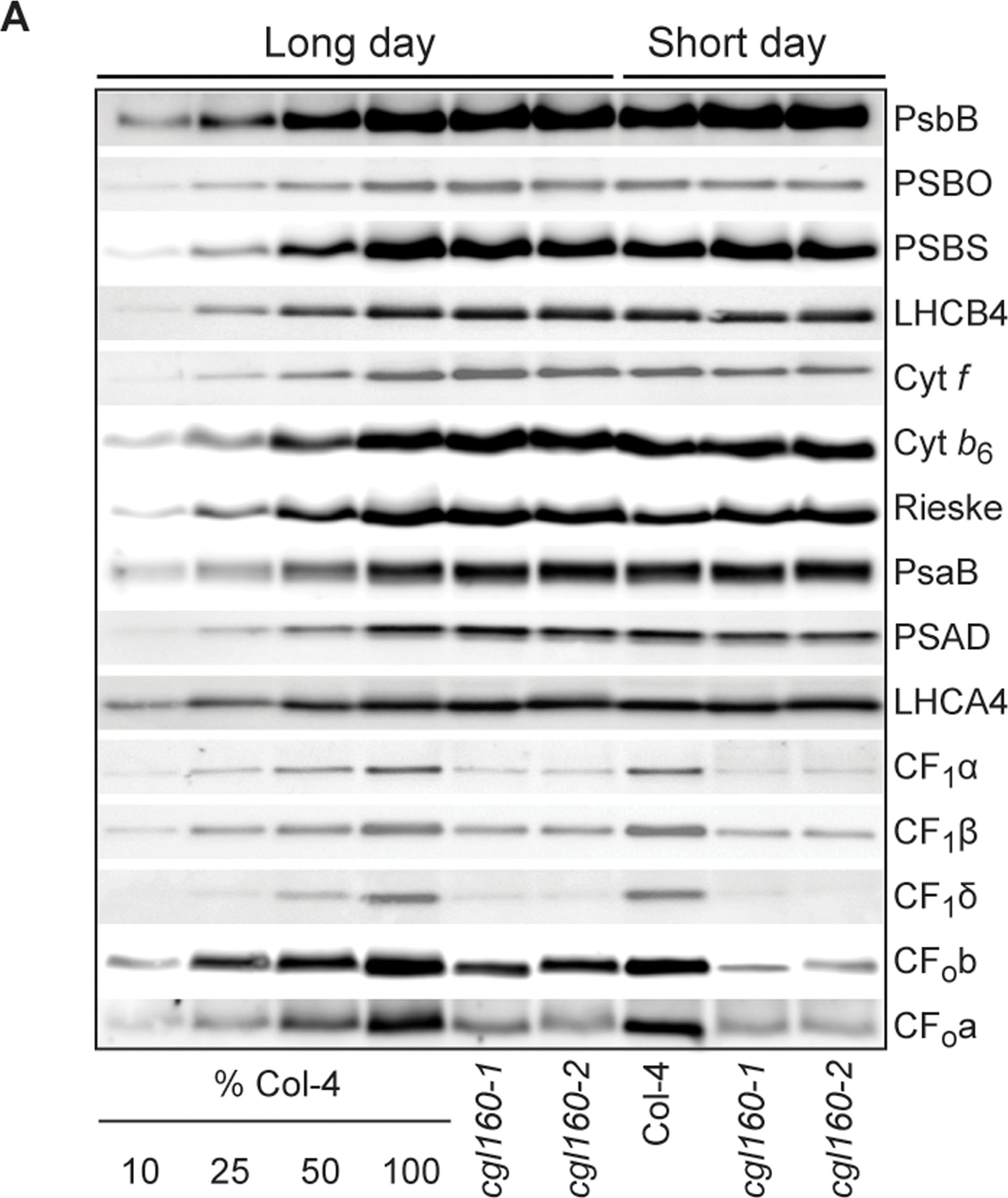
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 25835989
Journal: PLoS One
Figure Number: 7A
Published Date: 2015-04-04
First Author: Fristedt, R., Martins, N. F., et al.
Impact Factor: 2.942
Open PublicationAltered protein accumulation and stability of the chloroplast ATP synthase in the cgl160 mutant visualized by immunoblotting.A. Immunoblots with antibodies against essential subunits of the photosynthetic protein complexes of wild-type (Col-4) Arabidopsis and the two cgl160 T-DNA insertion lines grown under long-day and short-day conditions. Isolated thylakoid membranes were used, and equal amounts of chlorophyll were loaded onto the SDS-PAGE gel. For approximate quantification, wild-type samples from long-day plants were diluted to 10%, 25% and 50%, respectively. Accumulation of PSII was probed with antibodies against PsbB and PSBO. Additionally, the PSBS protein involved in NPQ and the minor PSII antenna protein LHCB4 were probed. Accumulation of the cytochrome b6f complex was probed with antibodies against the essential subunits PetA (cytochrome f), PetB (cytochrome b6), and PETC (Rieske protein). Accumulation of PSI was probed with antibodies against the reaction center subunit PsaB and the stromal ridge subunit PsaD. ATP synthase accumulation was probed with antibodies against the CF1 subunits AtpA (CF1?), AtpB (CF1?) and AtpD (CF1?) and antibodies against the CF0 subunits AtpF (CF0b) and AtpI (CF0a). B. Loading difference estimation for immunoblotting CF1 between wild type and cgl160-1. To obtain similar immunoblotting signal three times more (15 ?g protein) was needed for cgl160-1 compared to wild type (5 ?g protein). C. Maintenance of CF1 was measured by incubating leaves from wild type and cgl160-1 in solution containing the plastid protein synthesis inhibitor chloramphenicol for the indicated time points. Protein extract was isolated and separated by SDS-PAGE, immunoblotted and probed with specific antibodies against CF1 and LHCB2.1. Three times more protein was loaded from the mutant to obtain equal level of CF1 immunoblotting signal, as specified in B.
Reactant: Nicotiana tabacum (Common tobacco)
Application: Western Blotting
Pudmed ID: 28180288
Journal: J Exp Bot
Figure Number: 5A
Published Date: 2017-02-01
First Author: Schöttler, M. A., Thiele, W., et al.
Impact Factor: 6.088
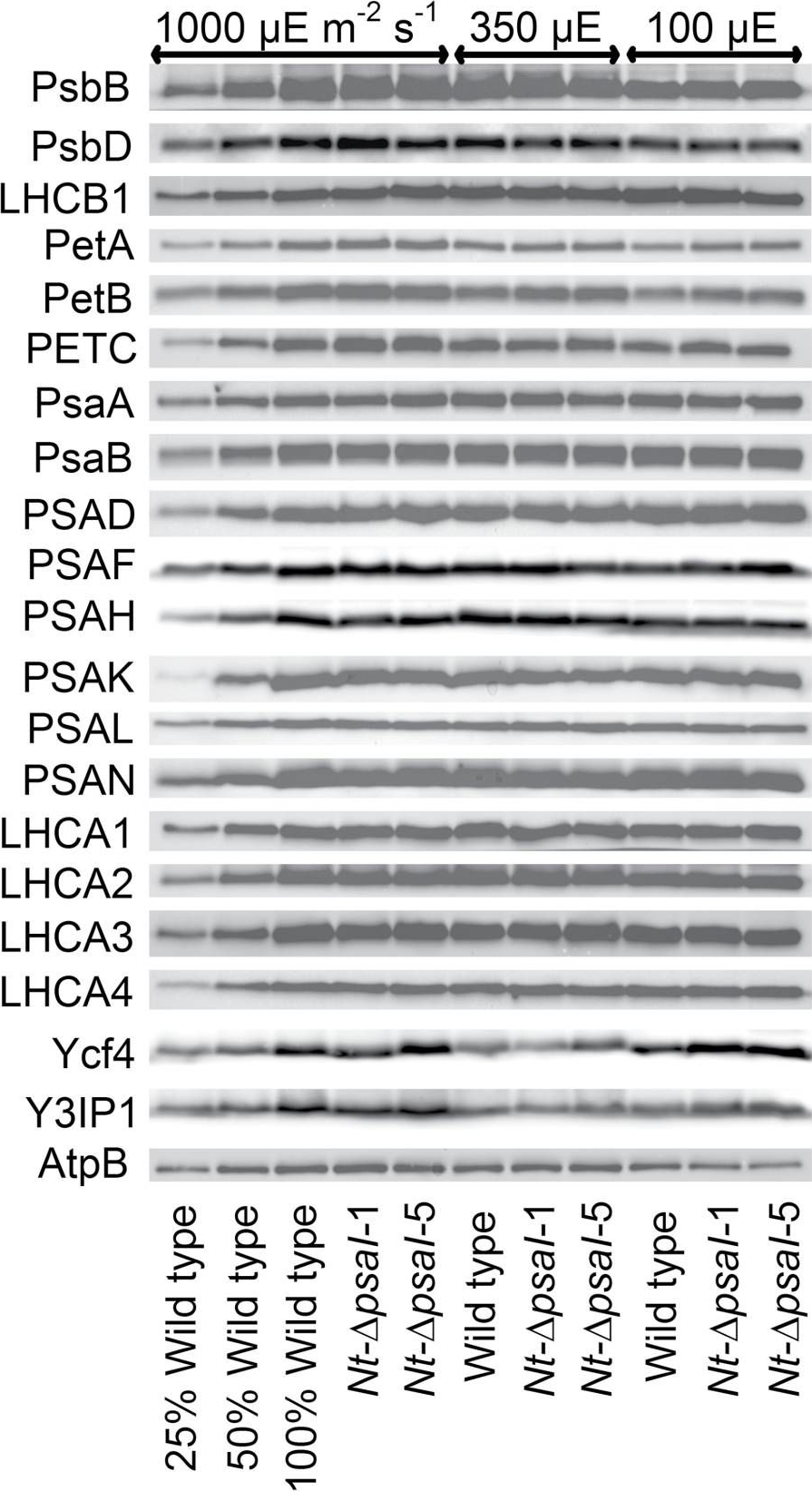
Open PublicationImmunoblot analysis of photosynthetic complex accumulation in wild-type tobacco and the two ?psaI lines grown under low, intermediate, and high-light conditions. Because the accumulation of most tested proteins was highest under high-light conditions, lanes one to three contain samples diluted to 25%, 50%, and a 100% sample of wild-type tobacco grown under high-light conditions, to allow for semi-quantitative determination of changes in protein abundance. Lanes four and five contain the two transplastomic lines grown at 1000 µE m?2 s?1. Lanes six to eight contain wild-type tobacco and the mutants grown at intermediate light intensities, and lanes nine to eleven contain samples grown at low light intensities. For PSII, the accumulation of the essential subunits PsbB (CP43) and PsbD (D2) and the LHCB1 antenna protein were determined, while for the cytochrome b6f complex, the accumulation of the essential redox-active subunits PetA (cytochrome f), PetB (cytochrome b6), and PETC (Rieske FeS protein) was tested. AtpB was probed as an essential subunit of the chloroplast ATP. For PSI, in addition to the three essential plastome-encoded subunits PsaA, PsaB, and PsaC, the accumulation of the nuclear-encoded subunits PSAD, PSAH, PSAK, PSAL, and PSAN and of the four LHCI proteins (LHCA1, LHCA2, LHCA3, LHCA4) was determined. Finally, we examined the accumulation of Ycf4, the chloroplast-encoded PSI-biogenesis factor encoded in the same operon as PsaI, and the nuclear-encoded assembly factor Y3IP1.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 28791032
Journal: Front Plant Sci
Figure Number: 2A
Published Date: 2017-08-10
First Author: Kohzuma, K., Froehlich, J. E., et al.
Impact Factor: 5.435
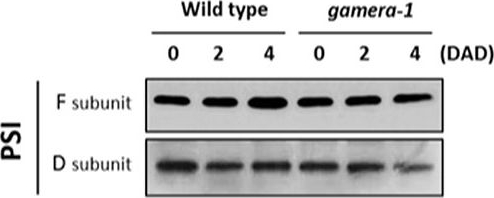
Open PublicationChanges in the protein levels of photosynthetic components under extended dark exposure in wild-type and gamera-1. Immunoblot detection of photosynthetic proteins from leaves of Ws and gamera-1 plants incubated after dark adaptation for 0, 2, and 4 days was examined. Specifically, essentially thylakoid fractions were assayed to determine the content of the following proteins: ?-subunit of ATP synthase; the D1 protein, OEC17, OEC23, and OEC33 of PSII; Cyt f and Rieske protein of the cytochrome b6f complex; and the F and D subunits of PSI, after extended dark treatment. Proteins were resolved via SDS-PAGE gel based on equal microgram chlorophyll per lane loading and processed as described in Section “Materials and Methods”. The Large subunit of RuBisco and LHCII stained with either CBB or Ponceau red, respectively, are presented here as loading controls. DAD indicates days after dark adaptation.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 32269582
Journal: Front Plant Sci
Figure Number: 7B
Published Date: 2020-04-10
First Author: Pralon, T., Collombat, J., et al.
Impact Factor: 5.435
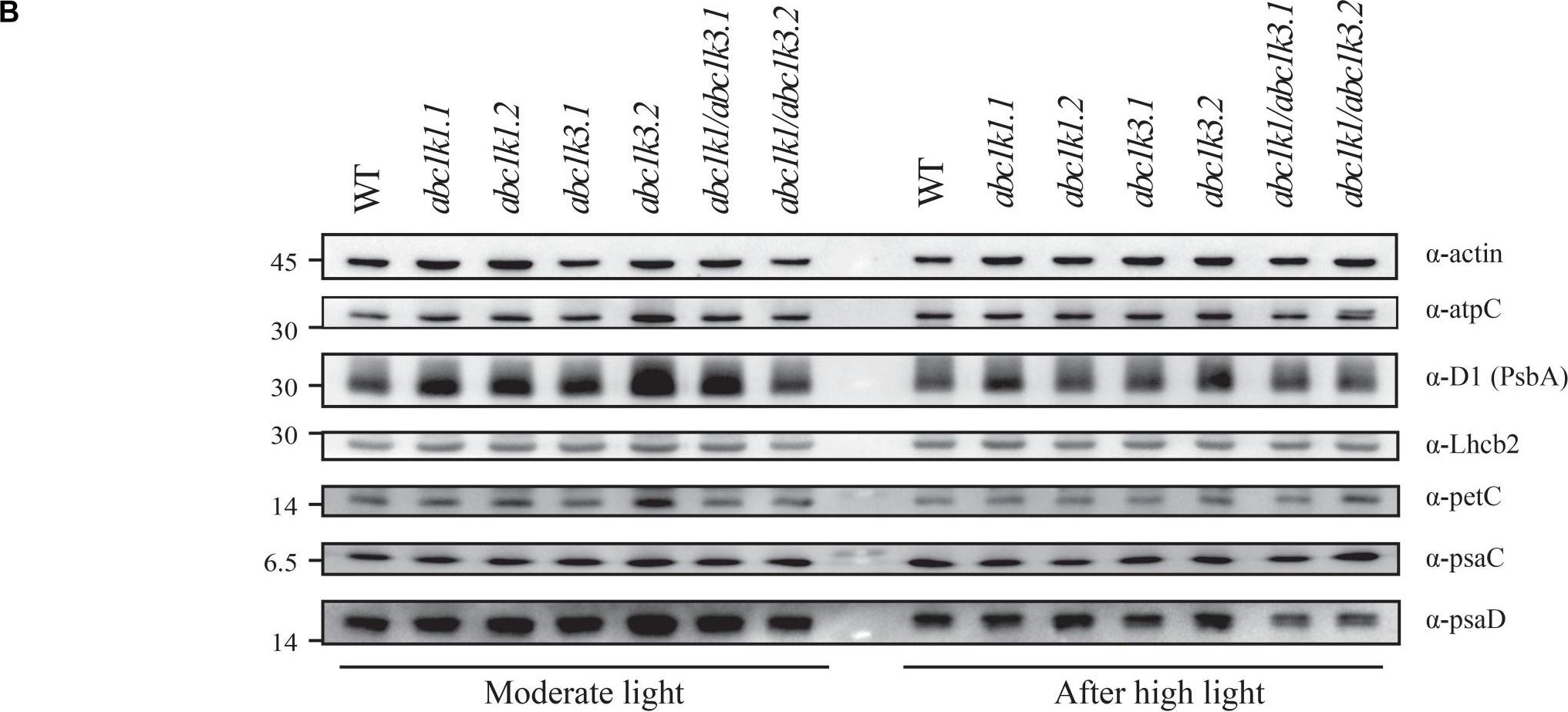
Open PublicationDouble mutant maintains thylakoid protein phosphorylation and state transitions after high light. (A) Total protein extracts of wild type (WT), abc1k1.1, -2, abc1k3.1, -2, and abc1k1/abc1k3.1, -2 light-exposed leaves were separated by SDS PAGE, transferred on nitrocellulose membrane and decorated with anti-phosphothreonine antibody. The main thylakoid phospho-proteins are indicated on the right according to their size. Core photosystem II proteins D1 (PsbA) and D2 (PsbD) are indicated together due their poor resolution. (B) The accumulation of the principal photosynthetic complexes was assessed using antibodies against specific subunits of each complex: anti-Lhcb2 for the major LHCII, anti-D1 (PsbA) for PSII, anti-PetC for cytochrome b6f, anti-PsaD and anti-PsaC for PSI, and anti-AtpC for ATP synthase. Actin signal is shown as a loading control. (C) Fluorescence quenching related to the state transitions (qT) of wild type (WT), abc1k1.1, -2, abc1k3.1, -2, and abc1k1/abc1k3.1, -2 under moderate light (120 ?mol of photons m–2 s–1) (ML) and after 3 h of high light (500 ?mol of photons m–2 s–1) (HL). qT was calculated from the maximal chlorophyll fluorescence measured after 10 min exposure to red light (660 nm) supplemented with far-red illumination (720 nm) “State 1” (FMST1) or to pure red light “State 2” (FMST2). Quenching related to state transition was calculated as qT = (FMST1 – FMST2)/FM. Each value represents the average of a pot containing 2–3 plants. Superscript letters are used to indicate statistically different groups (p < 0.05) by paired Student’s t-test.
Reactant: Synechocystis
Application: Western Blotting
Pudmed ID: 33383642
Journal: Life (Basel)
Figure Number: 1A
Published Date: 2020-12-29
First Author: Ka?a, R., Steinbach, G., et al.
Impact Factor: None
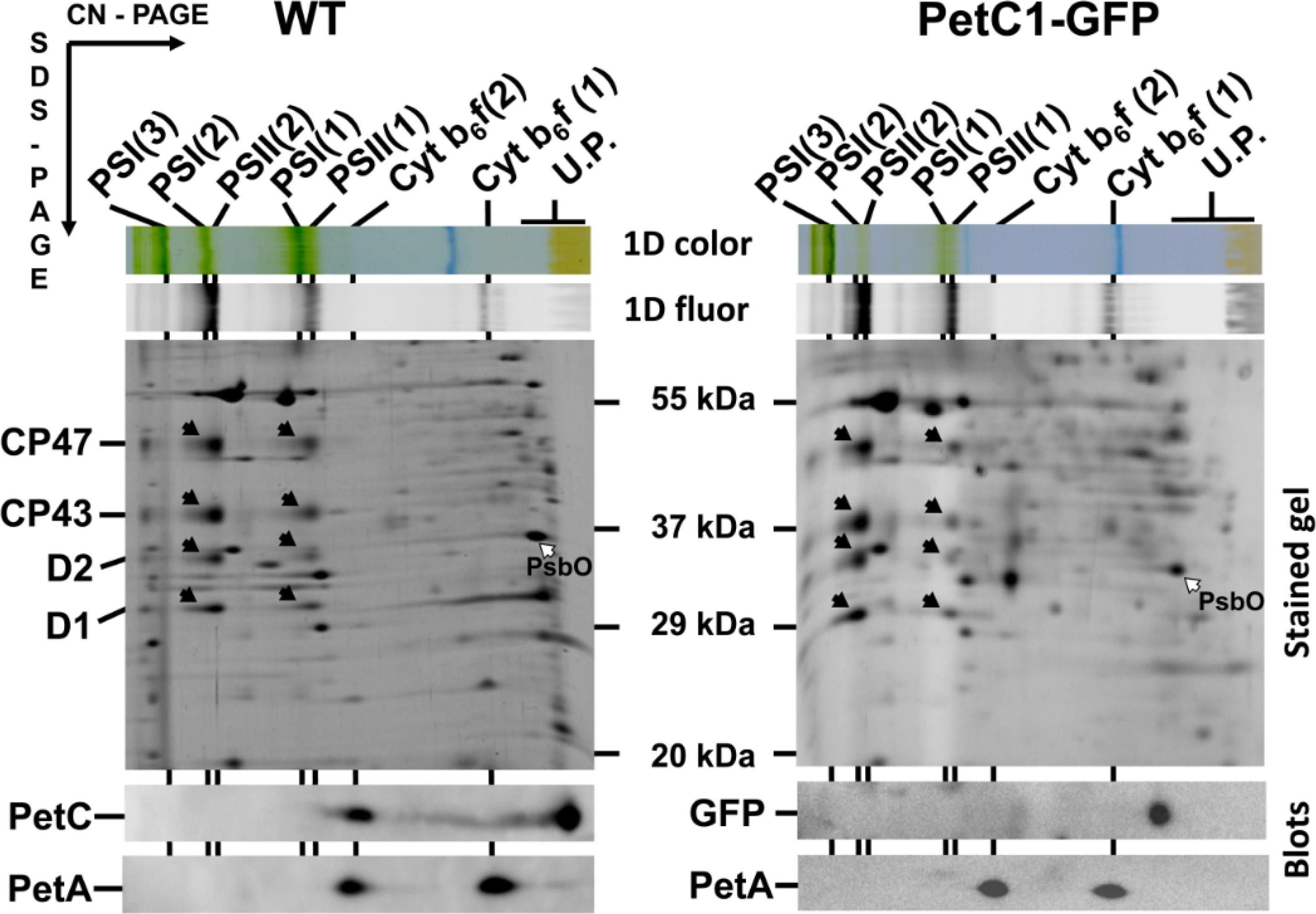
Open Publication2D analysis of membrane proteins of WT and GFP-PetC1 expressing strains. Membranes isolated from cells were analyzed by 2D CN/SDS-PAGE in combination with immunoblotting. Dimeric and monomeric Cyt b6-f complexes were identified using anti PetA antibody, PetC in WT was identified using general anti PetC antibody. Designation of complexes: PSI(3), PSI(2), PSI(1), trimeric, dimeric and monomeric PSI complexes, resp.; PSII(2) and PSII(1), dimeric and monomeric PSII core complexes, resp.; Cyt b6-f(2) and Cyt b6-f(1), dimeric and monomeric cytochrome b6-f complexes, resp.; U.P. unassembled proteins. The large PSII proteins CP47, CP43, D2 and D1 within PSII monomers and dimers (black arrows) and the unassembled PsbO (empty arrow) were identified previously by mass spectrometry [40,41] and by immunoblotting [42]. Each loaded sample contained 5 µg of CHL.
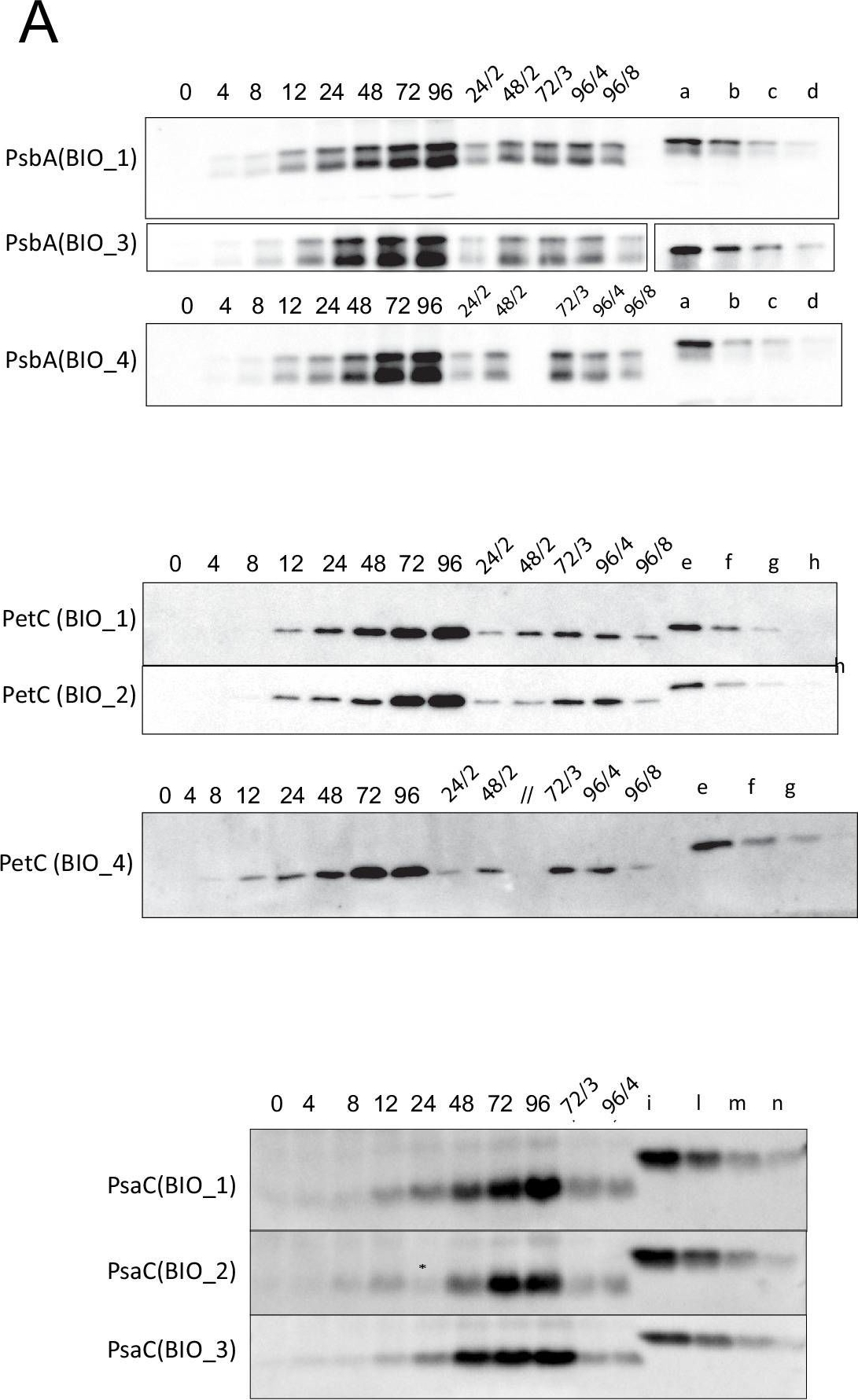
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 33629953
Journal: Elife
Figure Number: 6A
Published Date: 2021-02-25
First Author: Pipitone, R., Eicke, S., et al.
Impact Factor: 7.448
Open PublicationQuantification of photosynthesis-related proteins.(A) Immunodetection of PsbA, PetC, and PsaC during de-etiolation. Dilutions were used for the later time points to avoid saturation of the signal. (B) Different bands were detected by Amersham Imager program and quantified by Image QuantTL (Amersham). (C) Calibration curves were created using recombinant proteins (Agrisera). Calibration curve composition: PsbA 10 ng (A; lane a), 5 ng (b), 2.5 ng (c), and 1.25 ng (d); PetC 10 ng (e), 5 ng (f), 2.5 ng (g), and 1.25 ng (h); PsaC 3 ng (i), 1.5 ng (l), 0.75 ng (m), and 0.325 ng (n). Data indicate mean ± SD (n = 3–4). Raw data and calculations are shown in Figure 6—source data 1.
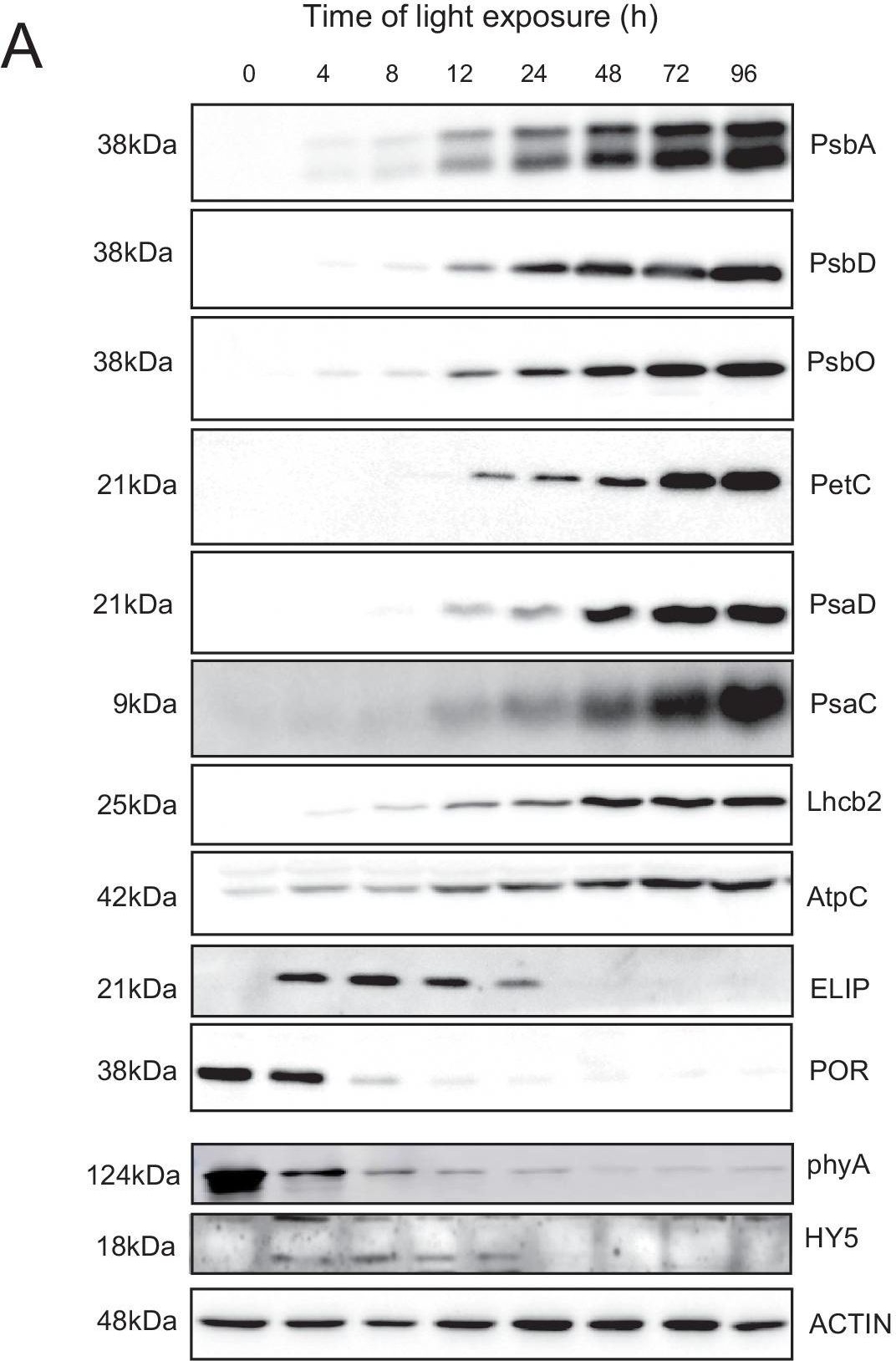
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 33629953
Journal: Elife
Figure Number: 6A
Published Date: 2021-02-25
First Author: Pipitone, R., Eicke, S., et al.
Impact Factor: 7.448
Open PublicationAccumulation dynamics of photosynthesis-related proteins during de-etiolation.Three-day-old etiolated seedlings of Arabidopsis thaliana were illuminated for 0 hr (T0), 4 hr (T4), 8 hr (T8), 12 hr (T12), 24 hr (T24), 48 hr (T48), 72 hr (T72), and 96 hr (T96) under white light (40 µmol/m2/s). (A) Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membrane and immunodetected with antibodies against PsbA, PsbD, PsbO, PetC, PsaD, PsaC, Lhcb2, AtpC, ELIP, POR, phyA, HY5, and ACTIN proteins. (B–C) Quantification of PsbA, PetC, and PsaC during de-etiolation. Heatmap (B) was generated after normalization of the amount of each protein relative to the last time point (T96). Graph (C) corresponds to the absolute quantification of proteins at T96. Error bars indicate ± SD (n = 3). Quantification of photosystem-related proteins during de-etiolation is detailed in Figure 6—figure supplement 1.Figure 6—source data 1.Quantitative data for immunoblot analysis.Quantitative data for immunoblot analysis.Quantification of photosynthesis-related proteins.(A) Immunodetection of PsbA, PetC, and PsaC during de-etiolation. Dilutions were used for the later time points to avoid saturation of the signal. (B) Different bands were detected by Amersham Imager program and quantified by Image QuantTL (Amersham). (C) Calibration curves were created using recombinant proteins (Agrisera). Calibration curve composition: PsbA 10 ng (A; lane a), 5 ng (b), 2.5 ng (c), and 1.25 ng (d); PetC 10 ng (e), 5 ng (f), 2.5 ng (g), and 1.25 ng (h); PsaC 3 ng (i), 1.5 ng (l), 0.75 ng (m), and 0.325 ng (n). Data indicate mean ± SD (n = 3–4). Raw data and calculations are shown in Figure 6—source data 1.
- Additional Information
-
Additional information: This product can be sold containing Proclin if requested.
- Background
-
Background: Rieske Iron-Sulfur Protein (Q9ZR03) is located in chloroplast thylakoid membrane as a component of cytochrome b6-f complex, which mediates electron transfer between photosystem II (PSII) and photosystem I (PSI), cyclic electron flow around PSI, and state transitions. Alternative names: Rieske iron-sulfur protein, RISP, ISP, plastohydroquinone:plastocyanin oxidoreductase iron-sulfur protein, proton gradient regulation protein 1
- Product Citations
-
Selected references: Collombat et al. (2025). Arabidopsis conditional photosynthesis mutants abc1k1 and var2 accumulate partially processed thylakoid preproteins and are defective in chloroplast biogenesis. Commun Biol . 2025 Jan 22;8(1):111. doi: 10.1038/s42003-025-07497-y.Krynicka, et al. (2023) FtsH4 protease controls biogenesis of the PSII complex by dual regulation of high light-inducible proteins. Plant Commun. 2023;4(1):100502. doi:10.1016/j.xplc.2022.100504
Penzler et al. (2024). A pgr5 suppressor screen uncovers two distinct suppression mechanisms and links cytochrome b6f complex stability to PGR5. Plant Cell. 2024 Mar 27:koae098. doi: 10.1093/plcell/koae098.
Ermakova et al. (2024). Chloroplast NADH dehydrogenase-like complex-mediated cyclic electron flow is the main electron transport route in C4 bundle sheath cells. New Phytol. 2024 Jul 22.doi: 10.1111/nph.19982.
Ermakova et al. (2022) Enhanced abundance and activity of the chloroplast ATP synthase in rice through the overexpression of the AtpD subunit. J Exp Bot. 2022 Jul 29:erac320. doi: 10.1093/jxb/erac320. Epub ahead of print. PMID: 35904136.
Dai et al. (2023). Hypothetical chloroplast reading frame 51 encodes a photosystem I assembly factor in cyanobacteria. Plant Cell. 2023 Dec 26:koad330.doi: 10.1093/plcell/koad330.
Pipitone et al. (2021). A multifaceted analysis reveals two distinct phases of chloroplast biogenesis during de-etiolation in Arabidopsis. Elife. 2021 Feb 25;10:e62709. doi: 10.7554/eLife.62709. PMID: 33629953; PMCID: PMC7906606.
Kana et al. (2020). Fast Diffusion of the Unassembled PetC1-GFP Protein in the Cyanobacterial Thylakoid Membrane. Life (Basel). 2020 Dec 29;11(1):E15. doi: 10.3390/life11010015. PMID: 33383642.
Zhang et al. (2020). Enhanced Relative Electron Transport Rate Contributes To Increased Photosynthetic Capacity In Autotetraploid Pak Choi. Plant Cell Physiol. 2020 Jan 6. pii: pcz238. doi: 10.1093/pcp/pcz238.
Pralon et al. (2019). Plastoquinone homoeostasis by Arabidopsis proton gradient regulation 6 is essential for photosynthetic efficiency. Commun Biol. 2019 Jun 20;2:220. doi: 10.1038/s42003-019-0477-4.
Koochak et al. (2019). The structural and functional domains of plant thylakoid membranes. Plant J. 2019 Feb;97(3):412-429. doi: 10.1111/tpj.14127.
Liang et al. (2018). Thylakoid-Bound Polysomes and a Dynamin-Related Protein, FZL, Mediate Critical Stages of the Linear Chloroplast Biogenesis Program in Greening Arabidopsis Cotyledons. Plant Cell. 2018 Jul;30(7):1476-1495. doi: 10.1105/tpc.17.00972. Epub 2018 Jun 7.
Koochak et al. (2018). The structural and functional domains of plant thylakoid membranes. Plant J. 2018 Oct 12. doi: 10.1111/tpj.14127.(Blue Native PAGE)
Du et al. (2018). Galactoglycerolipid Lipase PGD1 Is Involved in Thylakoid Membrane Remodeling in Response to Adverse Environmental Conditions in Chlamydomonas. Plant Cell. 2018 Feb;30(2):447-465. doi: 10.1105/tpc.17.00446.
Wood et al. (2018). Dynamic thylakoid stacking regulates the balance between linear and cyclic photosynthetic electron transfer. Nat Plants. 2018 Feb;4(2):116-127. doi: 10.1038/s41477-017-0092-7.
Schottler et al. (2017). The plastid-encoded PsaI subunit stabilizes photosystem I during leaf senescence in tobacco. J Exp Bot. 2017 Feb 1;68(5):1137-1155. doi: 10.1093/jxb/erx009.
Xing et al. (2017). Deletion of CGLD1 Impairs PSII and Increases Singlet Oxygen Tolerance of Green Alga Chlamydomonas reinhardtii. Front. Plant Sci., 15 December 2017.
Zang et al. (2017). Characterization of the sulfur-formation (suf) genes in Synechocystis sp. PCC 6803 under photoautotrophic and heterotrophic growth conditions. Planta. 2017 Jul 14. doi: 10.1007/s00425-017-2738-0.
Nath et al. (2016). A Nitrogen-Fixing Subunit Essential for Accumulating 4Fe-4S-Containing Photosystem I Core Proteins. Plant Physiol. 2016 Dec;172(4):2459-2470. Epub 2016 Oct 26.
Zhang et al. (2016). A new paradigm for producing astaxanthin from the unicellular green alga Haematococcus pluvialis. Biotechnol Bioeng. 2016 Oct;113(10):2088-99. doi: 10.1002/bit.25976. Epub 2016 Mar 28.
Fristedt et al. (2015). The thylakoid membrane protein CGL160 supports CF1CF0 ATP synthase accumulation in Arabidopsis thaliana. PLoS One. 2015 Apr 2;10(4):e0121658. doi: 10.1371/journal.pone.0121658. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
Agrisera Educational Posters CollectionRubisco quantitation in plant and algal samples using Agrisera anti-petC global antibody and PetC protein standard
Methodology: Plant samples are generally ground with liquid nitrogen in a mortar and pestle. The resulting powder is transferred to a plastic tube. Algal samples can be either concentrated by centrifugation or, preferably, by filtration onto glass fiber filters. Solubilization is performed in Agrisera protein extraction buffer (PEB, AS08 300) containing 0.1mg/mL PefaBloc SC (AEBSF) protease inhibitor (Roche). Disruption is most optimally obtained through flash freezing of the sample in liquid nitrogen alternated with thawing by sonication with a microtip. This process can be repeated depending on the toughness of the sample. The sample is adjusted to 50 mM dithiothreitol and heated to 70°C for 5 minutes. Samples are cooled and centrifuged briefly prior to electrophoresis.
Optimal quantitation is achieved using moderate sample loads per gel lane, generally 0.5 to 2.5 ug total protein, depending on the abundance of the target protein.
Electrophoresis and Immunoblotting: Once solubilized, the proteins can be separated electrophoretically in a number of systems. We obtain optimal results with the Invitrogen NuPAGE gel system using Bis-Tris 4-12% gradient gels. Proteins are separated in MES SDS running buffer according to the manufacturer’s recommendations at 200 V for 35 minutes. The gels are transferred to PVDF in the same apparatus, the SureLock XCell blot module, for 60 minutes at 30 V for a single gel or 80 minutes for a pair. Following transfer the blots are blocked with non-fat dry milk up to 10 % in TBS-T, for 1 h/RT with gentle agitation. The blot is incubated with primary antibody, usually at 1:25 000 to 1:50 000, for 1 h/RT (if extreme femtogram detection reagents are used) or in lower primary antibody dilution for less sensitivie reagents (mid picogram and lower).
For quantitation a relatively high primary antibody: target protein ratio gives more reliable results than immunoblots at low ratios of primary antibody:target protein.
The blot is washed extensively in TBS-T (twice briefly, once for 15 minutes and three times for five minutes). The blot is incubated with secondary antibody, for example goat anti-rabbit IgG horse radish peroxidase conjugated, AS09 602 (Agrisera), for 1h/RT. The blot is washed as above and developed with ECL detection reagents.Quantitation: When quantitated standards are included on the blot, the samples can be quantitated using the available software. Excellent quantitation can be obtained with images captured on the Bio-Rad Fluor-S-Max or equivalent instrument using Bio-Rad QuantityOne software. The contour tool is used to select the area for quantitation and the values are background subtracted to give an adjusted volume in counts for each standard and sample. Using above protocol linear standard curves are generated over 1-1.5 orders of magnitude range in target load. It is important to note that immunodetections usually show a strongly sigmoidal signal to load response curve, with a region of trace detection of low loads, a pseudolinear range and a region of saturated response with high loads. For immunoquantitation it is critical that the target proteins in the samples and the standard curve fall within the pseudolinear range. Our total detection range using this protocol spans over 2 orders of magnitude, but the quantifiable range is narrower.
References:
MacKenzie et al (2005). Large reallocations of carbon, nitrogen and photosynthetic reductant among phycobilisomes, photosystems and Rubisco during light acclimation in Synechococcus elongatus are constrained in cells under low environmental inorganic carbon. Arch of Microbiol. 183: 190 - 202.
Recommended secondary antibodies: goat anti-rabbit HRP conjugated, goat anti-rabbit ALP conjugated
Bouchard et al. (2006) UVB effects on the photosystem II-D1 protein of phytoplankton and natural phytoplankton communities. Photochem and Photobiol 82: 936-951.
Morash et al. (2007) Macromolecular dynamics of the photosynthetic system over a seasonal developmental progression in Spartina alterniflora. Can J. of Bot. 85: 476-483(8)
Recommended chemiluminescent detection reagent: AgriseraECLBright - Reviews:
-
Sujith Puthiyaveetil | 2018-07-06A clear band around 22 kDa is visible with protein samples separated on a 11.5 % SDS-6M urea-PAGE gel. The primary antibody was diluted at 1:10000 in TBST. The samples tested were Arabidopsis and Craterostigma.Jerzy Kruk | 2013-11-21Analysis perfomed using Arabidopsis thylakoids, 4-20% SDS gradient gel, 2 ug chlorophyll per lane, AB dilution 1:10 000, the product observed at ca. 23 kDaHenning Strissel | 2008-12-05The antibody is producing a single band at the sice of 23 kDa. A 1:1000 dilution in TTBS with 5 % milk works perfect.