1
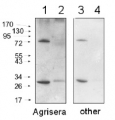
Anti-Lhcb2 | LHCII type II chlorophyll a/b-binding protein
AS01 003 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Photosynthetic eukaryotes including A. thaliana, A. hypogaea, B. sylvaticum, , C. arietinum, C. sinensis, C. quitensis Kunt Bartl, C. sativa, H. vulgare, C. reinhardtii, L. esculentum (Solanum lycopersicon), M. crystallinum, N. tabacum, O. sativa, P. patens, P. sativum, P. vulgaris, S. alba, S. oleracea, T. aestivum, Triticale, Z. mays
- Product Info
-
Immunogen: BSA-conjugated synthetic peptide derived from a highly conserved sequence of Lhcb2 proteins from angiosperms (monocots and dicots) and gymnosperms, including Arabidopsis thaliana Lhcb2.1 UniProt: Q9SHR7, TAIR: AT2G05100, Lhcb2.2 UniProt: Q9S7J7, TAIR:AT2G05070, Lhcb2.3 UniProt:Q9XF87, TAIR:AT3G27690
Host: Rabbit Clonality: Polyclonal Purity: Immunogen affinity purified serum in PBS pH 7.4. Format: Lyophilized Quantity: 50 µg Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Immunoprecipitation (IP), ImmunoGold (IG), Western blot (WB) Recommended dilution: 5 µl of antibody solution (IP), 1: 100 (IG), 1: 500 - 1 : 5000 (WB) Expected | apparent MW: 28.6 | 25 kDa for Arabidopsis thaliana
- Reactivity
-
Confirmed reactivity: Acer pseudoplatanus, Arabidopsis thaliana, Arachis hypogaea, Brachypodium sylvaticum, Camelina sinensis, Cicer arietum, Chlorella vulgaris, Colobanthus quitensis Kunt Bartl, Chlamydomonas reinhardtii, Cucumis sativus, Cytisus cantabricus (Wilk.) Rchb. F., Hieracium pilosella L., Hieracium pilosella L., Hordeum vulgare, Lasallia hispanica, Lycopersicon esculentum (Solanum lycopersicon), Miscanthus x giganteus, Mesembryanthemum crystallinum, Nicotiana benthamiana, Nicotiana tabacum, Oryza sativa, Pisum sativum, Phaseolus coccineus L., Phaseolus vulgaris, Physcomitrium patens, Setaria viridis, Sinapsis alba, Spinacia oleracea, Syntrichia muralis (Hedw.) Raab, Triticum aestivum, Triticale, Zea mays
Predicted reactivity: Algae, Dicots, Gymnosperms, Mosses Not reactive in: No confirmed exceptions from predicted reactivity are currently known - Application Examples
-

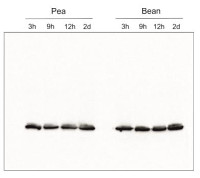
Species and variants: Pea – Pisum sativum L. Bean – Phaseolus coccineus L. 3h – 3 hours of cold exposure 9h – 9 hours of cold exposure 12h – 12 hours of cold exposure 2d – 2 days of cold exposure
Samples of isolated thylakoids containing 3 µg of chlorophyll were denatured with Laemmli buffer (1 vol : 1 vol) at 75 °C for 5 min. Denatured samples containing 1 µg of chlorophyll were loaded in the gel wells, separated on 12% SDS-PAGE gels and blotted for 45 min at 100 V to PVDF membrane using wet transfer. Blot was blocked with 5% milk in TBS-T for 60 min at room temperature (RT) with agitation. The blot was incubated with the primary antibody at a dilution of 1:500 in 1% Amersham™ ECL Prime Blocking Agent in TBS-T overnight at 4ºC with agitation. The antibody solution was decanted and the blot was washed 3 times for 5 min in TBS-T at RT with agitation. The blot was incubated using a secondary antibody (goat anti-rabbit IgG HRP conjugated, from Agrisera, AS09 602) diluted to 1: 25 000 in 1% milk in TBS-T for 1h at RT with agitation. The blot was washed 5 times for 5 min in TBS-T, 1 time for 5 min in TBS, 1 time for 5 min in 0.1 M Tris (pH 8.5), and developed for 4 min in substrates (0.188 mM coumaric acid, 1.25 mM luminol, 0.01% H2O2). Exposure time was 5 seconds in ChemiDoc scanner (BioRad).
Msc Małgorzata Krysiak, Faculty of Biology, University of Warsaw, Poland
Application examples: 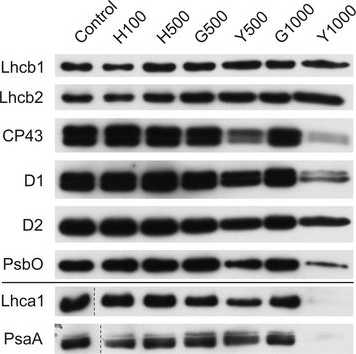
Reactant: Plant
Application: Western Blotting
Pudmed ID: 27590049
Journal: BMC Plant Biol
Figure Number: 9A
Published Date: 2016-09-02
First Author: Mazur, R., Sadowska, M., et al.
Impact Factor: 4.142
Open PublicationChanges of PSII and PSI antenna and core protein levels. Proteins from control and Tl-treated white mustard leaves were separated by SDS-PAGE followed by immunodetection with antibodies against Lhcb1, Lhcb2, Lhca1 (antenna proteins) and D1, D2, CP43, PsbO, PsaA (core proteins). Samples were loaded on the equal amount of chlorophyll (0.25 ?g). Description of samples abbreviation as given in the legend to Fig. 3
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 31240258
Journal: Commun Biol
Figure Number: 2b
Published Date: 2019-06-27
First Author: Pralon, T., Shanmugabalaji, V., et al.
Impact Factor: None
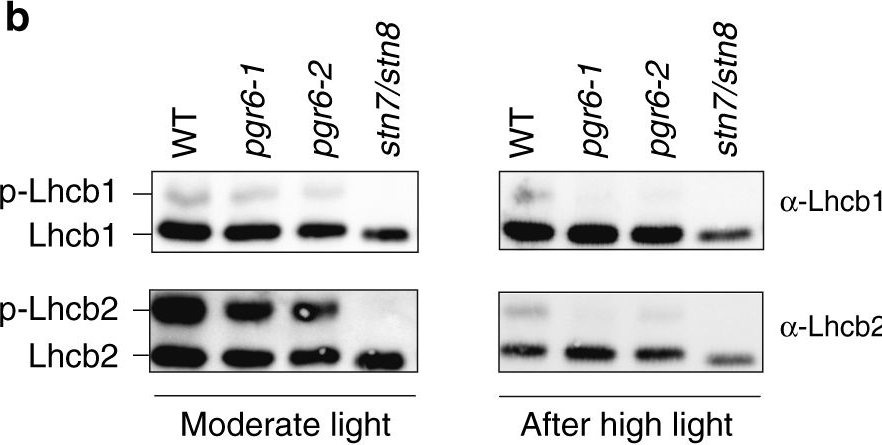
Open PublicationThylakoid protein phosphorylation and state transitions are disturbed after high light treatment in pgr6 background. a Total protein extracts of 4-week-old wild type (WT), pgr6-1, pgr6?2, sps2 and stn7/stn8 analysed by immunoblotting with anti-phosphothreonine antibody; the principal thylakoid phospho proteins are indicated on the right according to their size. Core photosystem II proteins D1 (PsbA) and D2 (PsbD) are indicated as a single band due to their poor resolution. Actin was used as a loading control. b Lhcb1 and Lhcb2 phosphorylation levels were visualised after separation on Phostag™-pendant acrylamide gels. The upper band corresponds to the phosphorylated form (p-), stn7/stn8 double mutant is a non-phosphorylated control. c Average transient of the variable room temperature chlorophyll fluorescence measured during the transition from red (660nm) supplemented with far-red light (720nm) state 1 to pure red light state 2 (n?=?4 independent pots containing 2–3 plants). The fluorescence curves from pgr6 and sps2 are shifted on the x-axis to allow visualising the FMST1 and FMST2 values. The x-axis time scale refers to the wild-type curve. d Calculated quenching related to state transition (qT?=?(FMST1?–?FMST2)/FM), expressed as the percentage of FM that is dissipated by the state 1 to state 2 transition, of wild type (WT), pgr6?1 and sps2 under moderate light (120?mol?m?2?s?1) (ML) and after 3?h of high light (500?mol?m?2?s?1) (HL). Whiskers and box plot shows the minimum, first quartile, median, average, third quartile and maximum of each dataset (n?=?4 biologically independent samples); p-values are calculated via a two-tailed Student’s t test. e STN7 phosphorylation level visualised after separation on Phostag™-pendant acrylamide gels. The upper band corresponds to the phosphorylated form (p-), a protein sample from stn7/stn8 double mutant was loaded as a control for the antibody specificity. Uncropped images of the membranes displayed in a, b and e are available as Supplementary Fig. 11. Data points for items c, d are available as Supplementary data 2
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 32003746
Journal: Elife
Figure Number: 8C
Published Date: 2020-01-31
First Author: Cazzonelli, C. I., Hou, X., et al.
Impact Factor: 7.448
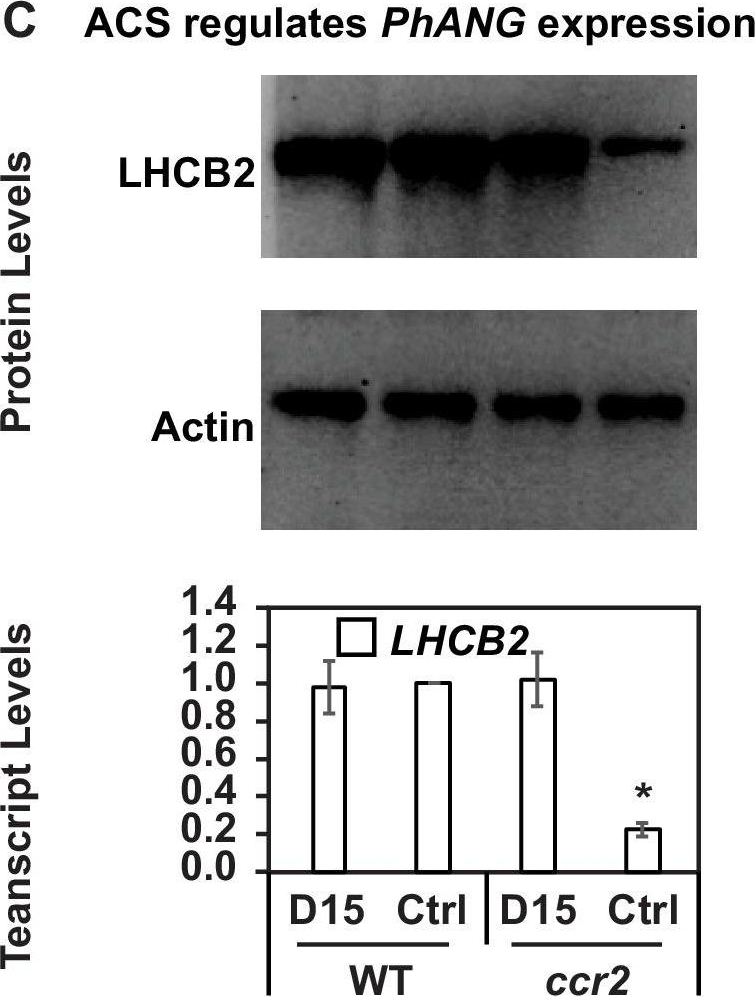
Open PublicationChemical inhibition of CCD activity revealed how a ccr2 generated apocarotenoid signal transcriptionally represses HY5 and LHCB2 expression during photomorphogenesis.(A) Transcript levels of PIF3 and HY5 in WT, ccr2, ccr2 det1-154 and det1-154 de-etiolated seedlings growing on MS media + /- D15. (B) Representative western blot images showing PIF3 and HY5 protein levels in WT, ccr2, ccr2 det1-154 and det1-154 de-etiolated seedlings growing on MS media + /- D15. The membrane was re-probed using anti-Actin antibody as an internal loading control. (C) Protein and transcript levels of LHCB2 expression in WT and ccr2 de-etiolated seedlings growing on MS media + /- D15. (D) Model showing how ACS regulates HY5 and LHCB2 expression in ccr2. Images of seedlings represent are cotyledons are coloured green or yellow to reflect the delay in chlorophyll biosynthesis induced by ACS as evidenced in Figure 6c. De-etiolation of seedlings was performed by transferring 4-d-old etiolated seedlings to continuous light for 3 d to induce photomorphogenesis. Statistical analysis denoted as a star was performed by pair-wise t-test (p<0.05). Error bars represent standard error of means. Ctrl; Control; Ctrl, D15; chemical inhibitor of CCD activity.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 32269582
Journal: Front Plant Sci
Figure Number: 7A,B
Published Date: 2020-04-10
First Author: Pralon, T., Collombat, J., et al.
Impact Factor: 5.435
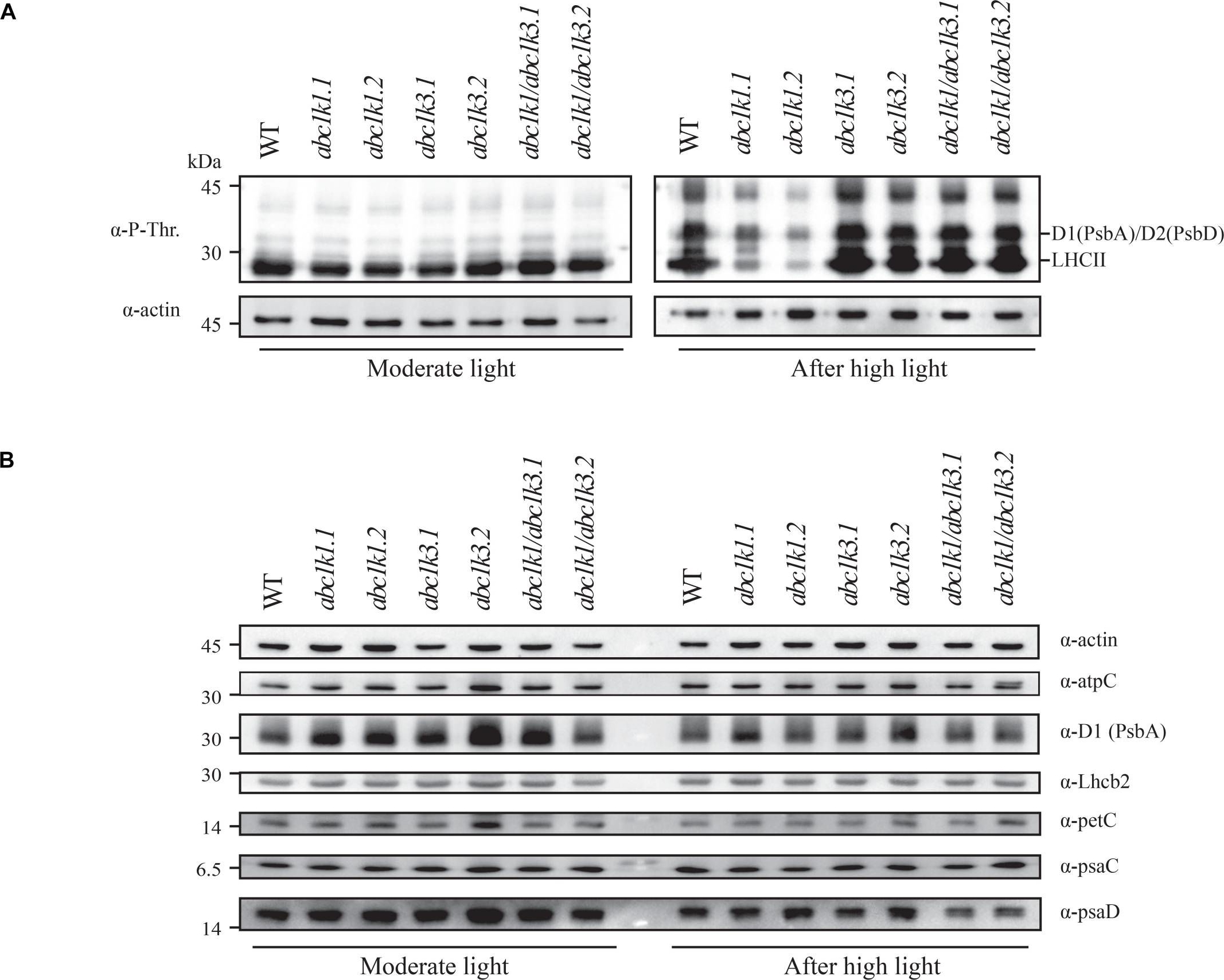
Open PublicationDouble mutant maintains thylakoid protein phosphorylation and state transitions after high light. (A) Total protein extracts of wild type (WT), abc1k1.1, -2, abc1k3.1, -2, and abc1k1/abc1k3.1, -2 light-exposed leaves were separated by SDS PAGE, transferred on nitrocellulose membrane and decorated with anti-phosphothreonine antibody. The main thylakoid phospho-proteins are indicated on the right according to their size. Core photosystem II proteins D1 (PsbA) and D2 (PsbD) are indicated together due their poor resolution. (B) The accumulation of the principal photosynthetic complexes was assessed using antibodies against specific subunits of each complex: anti-Lhcb2 for the major LHCII, anti-D1 (PsbA) for PSII, anti-PetC for cytochrome b6f, anti-PsaD and anti-PsaC for PSI, and anti-AtpC for ATP synthase. Actin signal is shown as a loading control. (C) Fluorescence quenching related to the state transitions (qT) of wild type (WT), abc1k1.1, -2, abc1k3.1, -2, and abc1k1/abc1k3.1, -2 under moderate light (120 ?mol of photons m–2 s–1) (ML) and after 3 h of high light (500 ?mol of photons m–2 s–1) (HL). qT was calculated from the maximal chlorophyll fluorescence measured after 10 min exposure to red light (660 nm) supplemented with far-red illumination (720 nm) “State 1” (FMST1) or to pure red light “State 2” (FMST2). Quenching related to state transition was calculated as qT = (FMST1 – FMST2)/FM. Each value represents the average of a pot containing 2–3 plants. Superscript letters are used to indicate statistically different groups (p < 0.05) by paired Student’s t-test.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 33322466
Journal: Biomolecules
Figure Number: 7A
Published Date: 2020-12-11
First Author: Andreeva, A. A., Vankova, R., et al.
Impact Factor: None
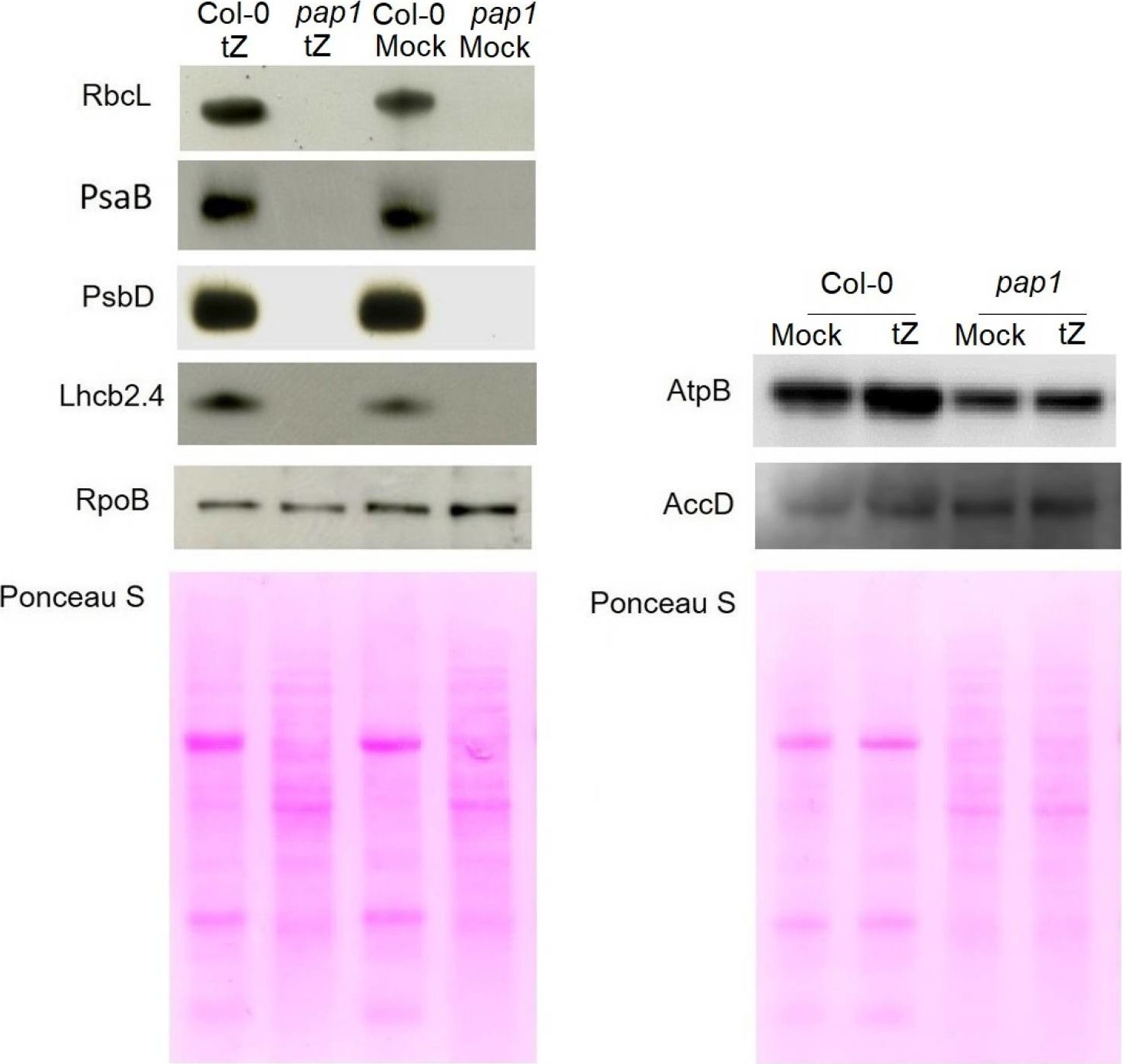
Open PublicationImmunoblot analysis of the photosynthetic proteins on the basis of equal total Ponceau S dye stained blot with proteins from leaves of wild type plants and pap1 mutant grown on MS medium in Petri dishes for four weeks under a 16 h light/8 h dark photoperiod at 23 °C with 100 ?E m?2 s?1. Proteins were visualized by immunoblotting using antibodies specific for RbcL, PsaB, PsbD, AtpB, RpoB, AccD and Lhcb2.4 proteins.
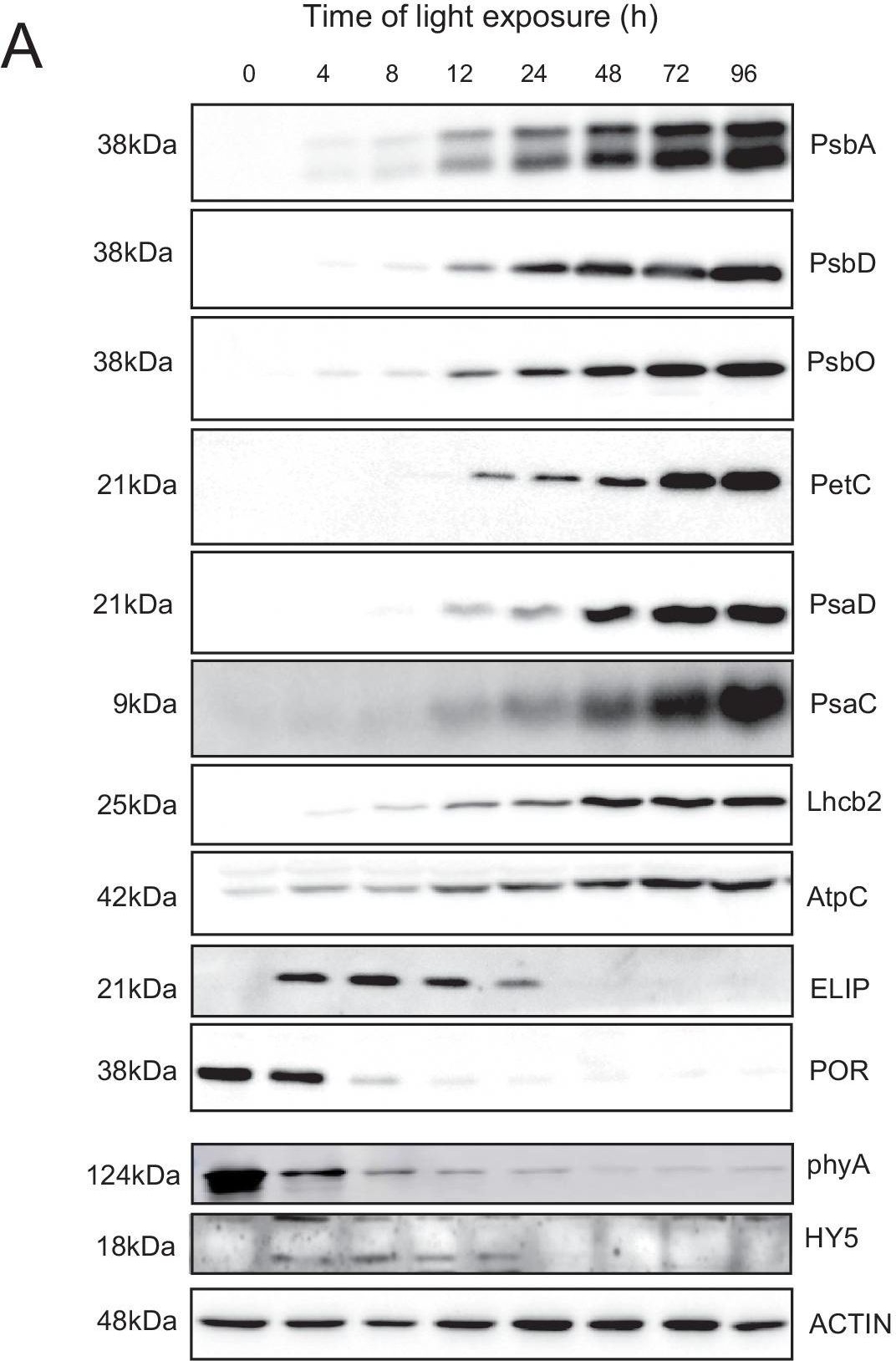
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 33629953
Journal: Elife
Figure Number: 6A
Published Date: 2021-02-25
First Author: Pipitone, R., Eicke, S., et al.
Impact Factor: 7.448
Open PublicationAccumulation dynamics of photosynthesis-related proteins during de-etiolation.Three-day-old etiolated seedlings of Arabidopsis thaliana were illuminated for 0 hr (T0), 4 hr (T4), 8 hr (T8), 12 hr (T12), 24 hr (T24), 48 hr (T48), 72 hr (T72), and 96 hr (T96) under white light (40 µmol/m2/s). (A) Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membrane and immunodetected with antibodies against PsbA, PsbD, PsbO, PetC, PsaD, PsaC, Lhcb2, AtpC, ELIP, POR, phyA, HY5, and ACTIN proteins. (B–C) Quantification of PsbA, PetC, and PsaC during de-etiolation. Heatmap (B) was generated after normalization of the amount of each protein relative to the last time point (T96). Graph (C) corresponds to the absolute quantification of proteins at T96. Error bars indicate ± SD (n = 3). Quantification of photosystem-related proteins during de-etiolation is detailed in Figure 6—figure supplement 1.Figure 6—source data 1.Quantitative data for immunoblot analysis.Quantitative data for immunoblot analysis.Quantification of photosynthesis-related proteins.(A) Immunodetection of PsbA, PetC, and PsaC during de-etiolation. Dilutions were used for the later time points to avoid saturation of the signal. (B) Different bands were detected by Amersham Imager program and quantified by Image QuantTL (Amersham). (C) Calibration curves were created using recombinant proteins (Agrisera). Calibration curve composition: PsbA 10 ng (A; lane a), 5 ng (b), 2.5 ng (c), and 1.25 ng (d); PetC 10 ng (e), 5 ng (f), 2.5 ng (g), and 1.25 ng (h); PsaC 3 ng (i), 1.5 ng (l), 0.75 ng (m), and 0.325 ng (n). Data indicate mean ± SD (n = 3–4). Raw data and calculations are shown in Figure 6—source data 1.
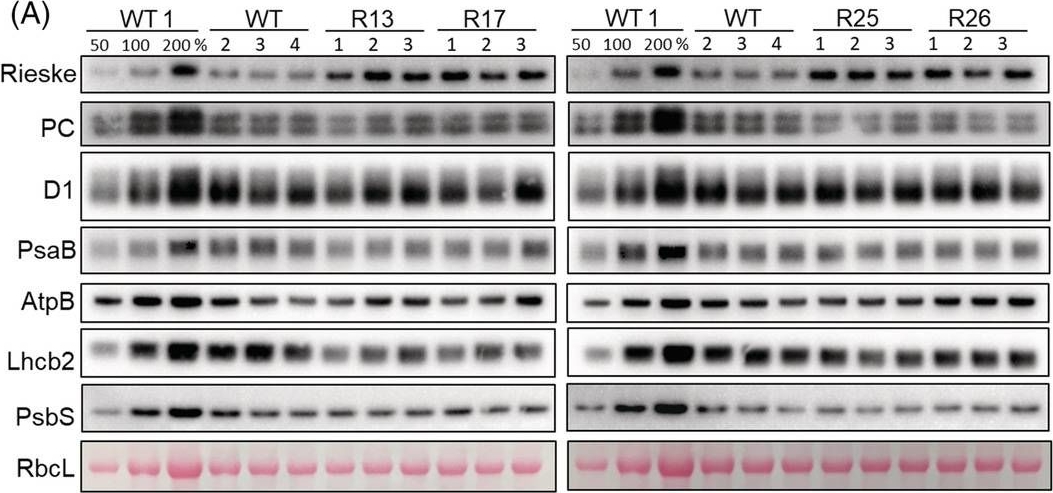
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 36259085
Journal: Physiol Plant
Figure Number: 4A
Published Date: 2022-11-01
First Author: Heyno, E.
Impact Factor: 4.812
Open PublicationAbundance of photosynthetic proteins in leaves of WT and Rieske‐OE plants. (A) Immunodetection of Rieske, plastocyanin (PC), D1 subunit of PSII, PsaB subunit of PSI, AtpB subunit of ATP synthase, Lhcb2 subunit of LHCII and PsbS in leaf protein extracts loaded on leaf area basis. The quantity of the large subunit of Rubisco (RbcL) was estimated from Ponceau stained membranes immediately after transfer. A titration series of one of the WT samples (WT1) was used for relative quantification. (B) Quantification of immunoblots relative to WT. mean ± SE, n = 4 biological replicates for WT, n = 3 for transgenic lines. Asterisks indicate statistically significant differences between transgenic lines and WT (Tukey test, *p < 0.05, **p < 0.01. ***p < 0.001).
- Additional Information
-
Additional information (application): Immunoprecipitation has been done using Immunoprecipitation kit from Roche, Cat.No. 11 719 386 001.
Protein is processed into mature form (Jansson 1999). - Background
-
Background: The major light-harvesting antenna complex II (LHCII) in photosynthetic eukaryotes is located in the thylakoid membrane of the chloroplast. It is a heterotrimeric complex formed by up to 3 different individual subtypes of chlorophyll a/b-binding proteins: Lhcb1, Lhcb2, and Lhcb3. Lhcb2 is often coded by several nuclear genes and is found together with Lhcb1 within the mobile LHCII trimers involved in state1-state2 transition.
A molecular characterisation of the LHCII proteins can be found in Caffarri et al. (2004) A Look within LHCII: Differential Analysis of the Lhcb1−3 Complexes Building the Major Trimeric Antenna Complex of Higher-Plant Photosynthesis. Biochemistry 43 (29): 9467–9476. - Product Citations
-
Selected references: Dziubek et al. (2025). Dissection of photosynthetic short and long-term acclimation to fluctuating light reveals specific functions within the chloroplast thioredoxin network. J Exp Bot . 2025 Mar 24:eraf121. doi: 10.1093/jxb/eraf121.
Ermakova et al. (2024). Chloroplast NADH dehydrogenase-like complex-mediated cyclic electron flow is the main electron transport route in C4 bundle sheath cells. New Phytol. 2024 Jul 22.doi: 10.1111/nph.19982.
Zhao et al. (2024). Psb28 protein is indispensable for stable accumulation of PSII core complexes in Arabidopsis.Plant J. 2024 May 26. doi: 10.1111/tpj.16844.
Ciesielska et al. (2024). S2P2-the chloroplast-located intramembrane protease and its impact on the stoichiometry and functioning of the photosynthetic apparatus of A. thaliana. Front Plant Sci. 2024 Mar 15:15:1372318. doi: 10.3389/fpls.2024.1372318.
Kim et al. (2024).Photoautotrophic cultivation of a Chlamydomonas reinhardtii mutant with zeaxanthin as the sole xanthophyll. Biotechnol Biofuels Bioprod. 2024 Mar 14;17(1):41. doi: 10.1186/s13068-024-02483-8.
Ye et al. (2023). The light-harvesting chlorophyll a/b-binding proteins of photosystem II family members are responsible for temperature sensitivity and leaf color phenotype in albino tea plant. J Adv Res . 2023 Dec 25:S2090-1232(23)00404-6.doi: 10.1016/j.jare.2023.12.017.
Singh, Muthamilarasan, Prasad (2022). SiHSFA2e regulated expression of SisHSP21.9 maintains chloroplast proteome integrity under high temperature stress. Cell Mol Life Sci. 2022;79(11):580. Published 2022 Nov 3. doi:10.1007/s00018-022-04611-10
Cazzaniga et al. (2022). Engineering astaxanthin accumulation reduces photoinhibition and increases biomass productivity under high light in Chlamydomonas reinhardtii. Biotechnol Biofuels Bioprod. 2022 Jul 11;15(1):77. doi: 10.1186/s13068-022-02173-3. PMID: 35820961; PMCID: PMC9277849.
Bru, Steen, Park, et al. (2022) The major trimeric antenna complexes serve as a site for qH-energy dissipation in plants. J Biol Chem. 2022;298(11):102519. doi:10.1016/j.jbc.2022.102521
Ivanov et al. (2022) The decreased PG content of pgp1 inhibits PSI photochemistry and limits reaction center and light-harvesting polypeptide accumulation in response to cold acclimation. Planta 255, 36 (2022). https://doi.org/10.1007/s00425-022-03819-0
Bychkov et al. (2022) The role of PAP4/FSD3 and PAP9/FSD2 in heat stress responses of chloroplast genes. Plant Sci. 2022 Sep;322:111359. doi: 10.1016/j.plantsci.2022.111359. Epub 2022 Jun 20. PMID: 35738478.
Lang et al. (2011).Simultaneous isolation of pure and intact chloroplasts and mitochondria from moss as the basis for sub-cellular proteomics. Plant Cell Rep. 2011 Feb;30(2):205-15.doi: 10.1007/s00299-010-0935-4. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
Agrisera Educational Posters Collection
- Reviews:
-
Annika Brünje | 2021-07-02I use this antibody regularly to compare Lhcb2 abundances in Arabidopsis during different stress treatments. It gives very nice signals in the immunodetection assay, even when only low amounts of total protein were loaded. I can definitely recommend.Soo Yeon Ko | 2020-11-23We always use this antiboy whenever we want to the this band in Western blot. The result is very good! I can recommend this antibody and the quality is also very very good :)Przemyslaw Kmiecik | 2017-02-01The lhcb2 antibody purchased from Agrisera is a high quality product. The antibody has high specificity - no background or unspecific bands. The resulting signal is really strong at short exposure times even when low concentration of the sample is used.Monica Colombo | 2015-01-30The antibody worked very well at 1:5000 in Arabidopsis with 3ug of proteinsMichele Grieco | 2014-06-16For this antibody I loaded an amount of sample with a chlorophyll concentration of only 0,5 µl/ml, and in western blotting analyses the ECL signal was very intense. Moreover, I did not get aspecific bands for any of these antibodies.Dorothee Staiger | 2013-08-28We used 1:5000 dilution - very strong bands with 10 micrograms of protein.Elke Stroeher | 2012-03-26Works for 2.5ug crude leaf protein extract (Arabidopsis thaliana) using a 1:5000 dilution. Specific: band at 25 kDa, and a much weaker band at 50kDa, probably a dimer.Maciej Garstka | 2009-03-19specific to pea, less to bean and spinach