1
Anti-HPR | Hydroxypyruvate reductase (peroxisomal matrix marker)
AS11 1797 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Arabidopsis thaliana, Pisum sativum
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide derived from known plant HRP sequences, including Arabidopsis thaliana UniProt: Q9C9W5,TAIR: At1g68010
Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Western blot (WB) Recommended dilution: 1 : 10 000 (WB) Expected | apparent MW: 43 kDa
- Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Pisum sativum Predicted reactivity: Chlamydomonas reinhardtii, Chlorella sp. ,Cucumis sativus, Glycine max, Oryza sativa, Populus albaxtremula, Ricinus communis, Volvox
Species of your interest not listed? Contact usNot reactive in: Nicotiana benthamiana, Nicotiana tabacum - Application Examples
-
Application example

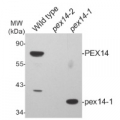
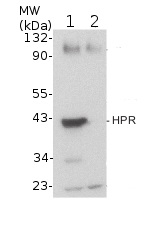
5-day-old light-grownwild-type (Columbia) (1) and hpr1-1 null mutant (SALK_143584) Arabidopsis thaliana seedlings were ground with a pestle in a 1.5 mL tube on dry ice (about 12 seedlings or enough to give ~ 20 µL of tissue), and double volume (~ 40 µL) of NuPAGE 2x loading buffer (Invitrogen) was added. After centrifugation, 20 µL of the supernatant was transferred to a fresh tube with 2.1 µL 0.5 M DTT and boiled at 100 °C for 5 minutes. Samples were loaded onto a NuPAGE 10% Bis-Tris gel (Invitrogen) next to Cruz Markers (Santa Cruz Biotechnology). After electrophoresis, proteins were transferred for 30 minutes at 24 V to a Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biotech) using NuPAGE transfer buffer (Invitrogen). The blot was blocked for 1 h at 4 °C in 8% non-fat dry milk in TBS-T (blocking buffer), and incubated overnight with agitation at 4˚C with primary antibodies, 1:10 000 diluted in blocking buffer. The antibody solution was decanted and the blot was rinsed twice for 5 min each at 4 °C in 8% non-fat dry milk in TBS-T with agitation. The blot was incubated with horseradish peroxidase-conjugated anti-rabbit secondary antibody (Santa Cruz Biotechnology) diluted to 1:5 000 in blocking buffer for 5 h at 4 °C with agitation. The blot was washed three times, for 5 min each, with TBS-T and developed with chemiluminescent detection reagent according to the manufacturer's instructions. Exposure time was 3 seconds.
Courtesy of Sarah Bukhart and Bonnie Bartel, Rice University, USA
Application examples: 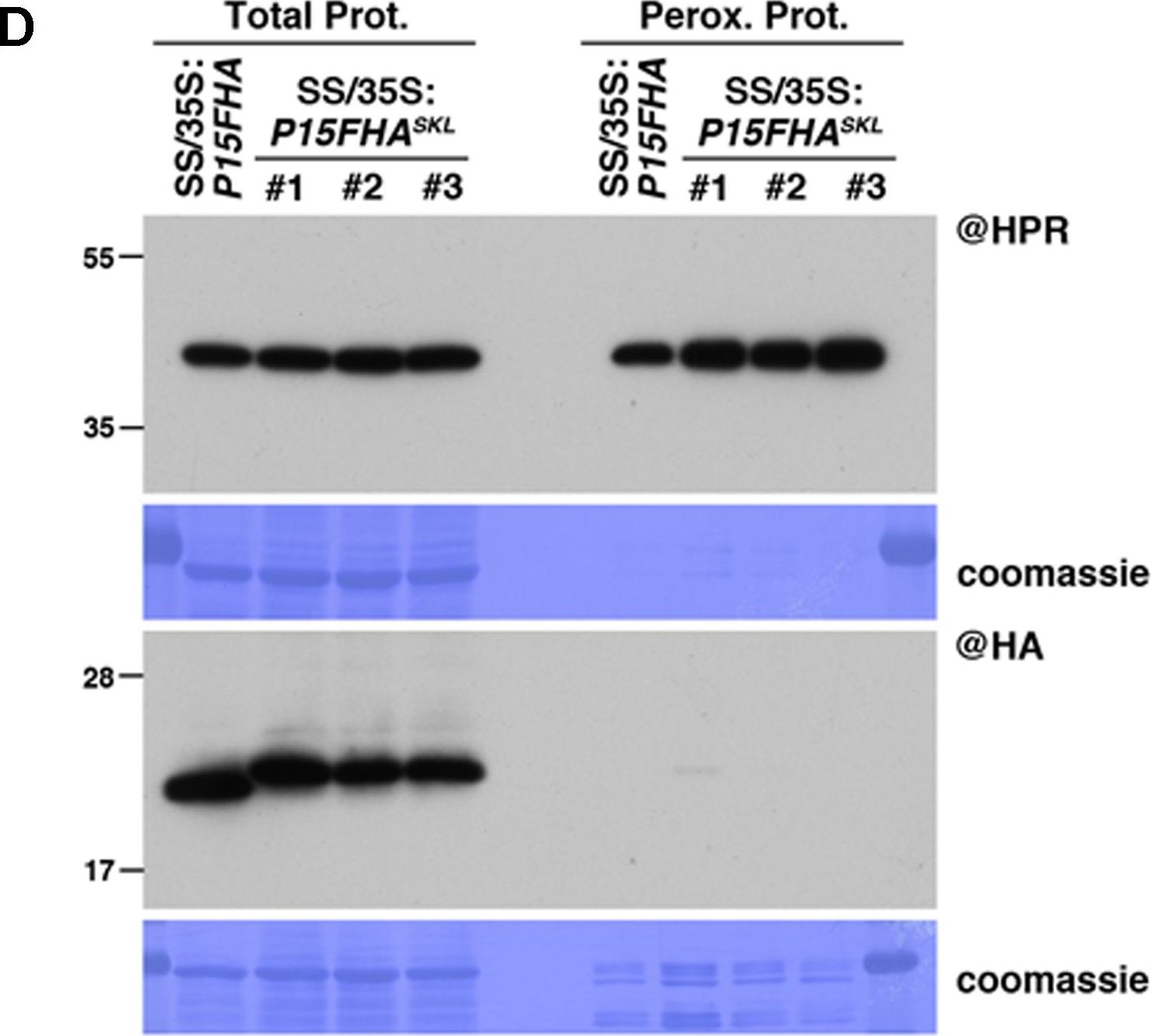
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 29479364
Journal: Front Plant Sci
Figure Number: 1D
Published Date: 2018-02-27
First Author: Incarbone, M., Ritzenthaler, C., et al.
Impact Factor: 5.435
Open PublicationPeroxisomal targeting of P15FHA reveals P15/small RNA interactions not detected by immunoprecipitation. (A) Northern analysis of small RNAs (SUL, IR71, REP2, miR159, miR160, miR160?, miR169, miR173) in total (left) and @HA immunoprecipitated (IP; right) fractions from SUC:SUL and 35S:P15FHA/SUC:SUL rosette leaves was obtained by sequential rounds of probing and stripping the same membrane. The three IP samples (#1, #2, #3) correspond to three technical replicates obtained from a pool of ten 35S:P15FHA/SUC:SUL plants. (B) Western analysis of P15FHA accumulation in total (left) and @HA IP (right) fractions obtained from the plants described in (A). (C) Northern analysis of small RNAs in total (left) and peroxisomal (perox.; right) fractions obtained from 35S:P15FHA/SUC:SUL and 35S:P15FHASKL/SUC:SUL plants. The small RNA species detected are the same as in (A). The three peroxisome samples (#1, #2, #3) correspond to three biological replicates of 35S:P15FHASKL/SUC:SUL plants. (D) Western analysis of plant peroxisomal marker hydroxypyruvate reductase (@HPR) and P15FHA/P15FHASKL (@HA) in total (left) and peroxisomal (right) fractions obtained from the plants described in (C). Note that more peroxisomal protein was loaded to detect P15FHA/P15FHASKL than to detect HPR. Northern analyses were performed on high-resolution gels. Accumulation of snU6 was used as RNA loading control, while coomassie staining was used as protein loading control. Figure source data can be found with the Supplementary information.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 29479364
Journal: Front Plant Sci
Figure Number: 2B
Published Date: 2018-02-27
First Author: Incarbone, M., Ritzenthaler, C., et al.
Impact Factor: 5.435
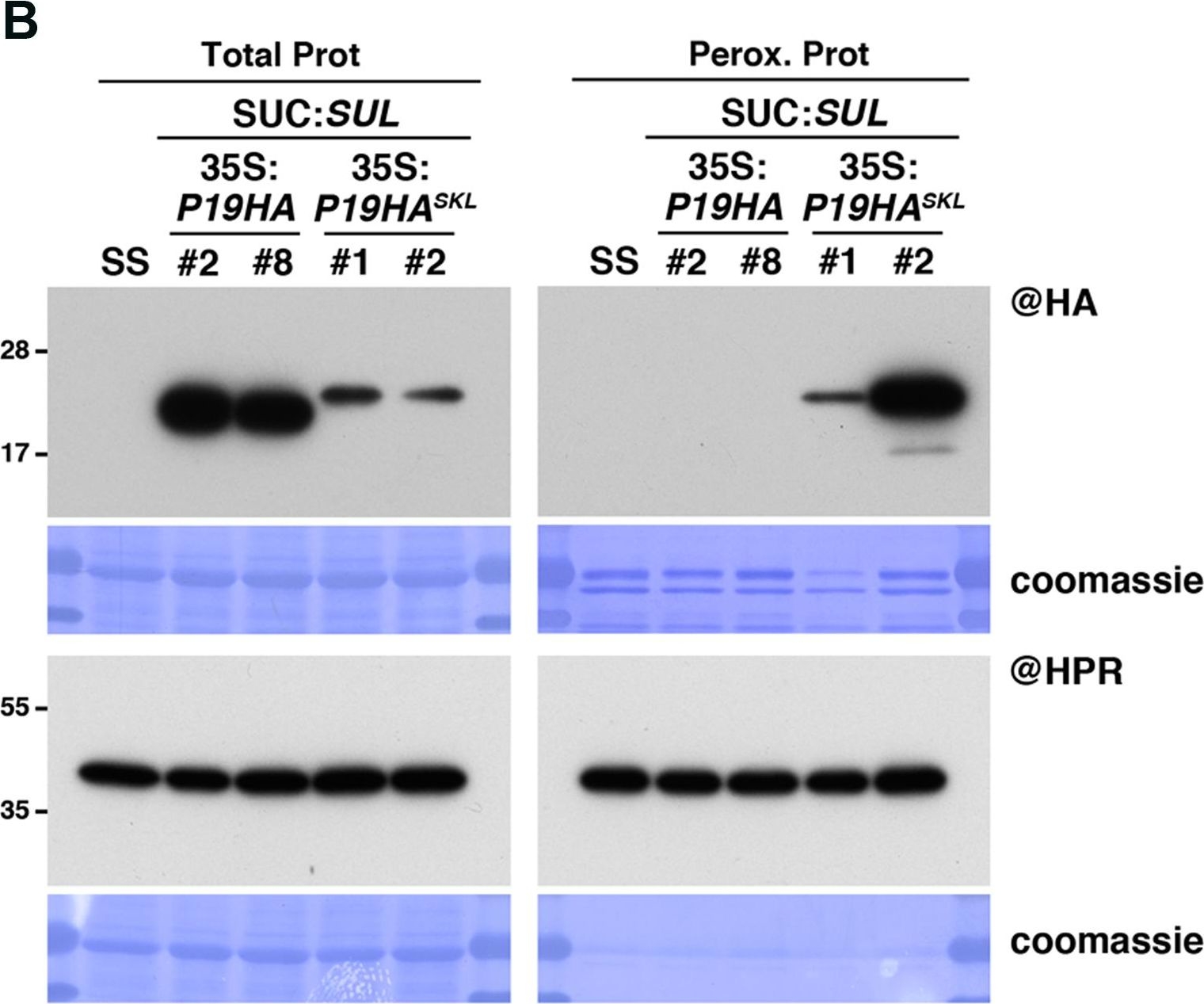
Open PublicationPeroxisomal targeting of P19HA leads to import of 21, 22, and 24nt small RNAs into these organelles. (A) Photos of SUC:SUL, 35S:P19HA/SUC:SUL (transgenic lines #2 and #8) and 35S:P19HASKL/SUC:SUL (transgenic lines #1 and #2) plants used for peroxisome isolation. (B) Western analysis of plant peroxisomal marker hydroxypyruvate reductase (@HPR) and P19HA/P19HASKL (@HA) in total (left) and peroxisomal (perox. right) fractions obtained from the plants described in (A). Note that more peroxisomal protein was loaded to detect P19HA/P19HASKL than to detect HPR. (C) Northern analysis of small RNAs (SUL, IR71, REP2, miR159) in total (left) and peroxisomal (right) fractions from the plants described in (A) was obtained by sequential rounds of probing and stripping the same membranes. Northern analysis was performed on a low-resolution gel. Accumulation of snU6 was used as RNA loading control, while coomassie staining was used as protein loading control. Figure source data can be found with the Supplementary Information.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 29479364
Journal: Front Plant Sci
Figure Number: 3D
Published Date: 2018-02-27
First Author: Incarbone, M., Ritzenthaler, C., et al.
Impact Factor: 5.435
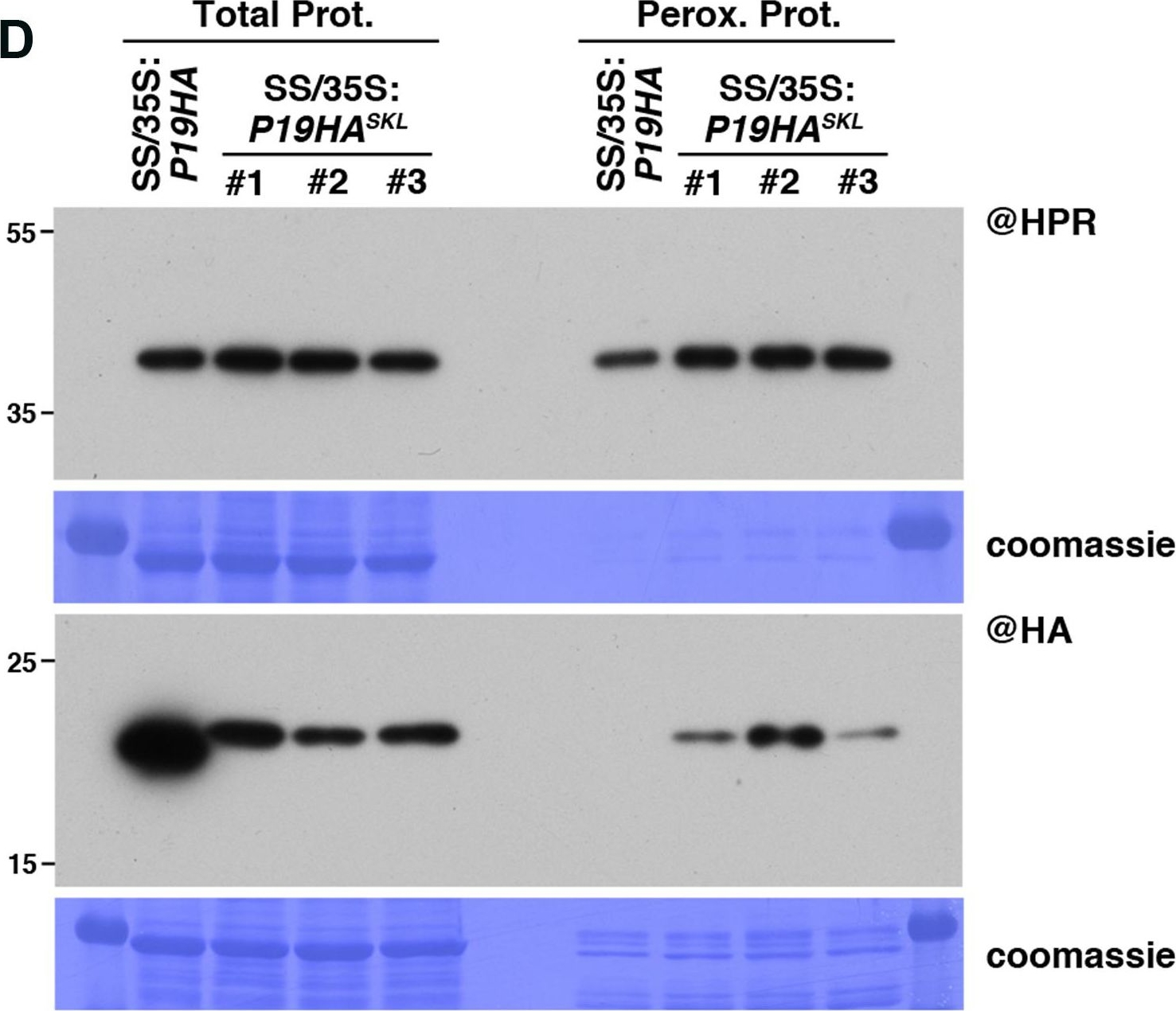
Open PublicationPeroxisomal targeting of P19HA reveals P19/small RNA interactions not detected by immunoprecipitation. (A) Northern analysis of small RNAs (SUL, IR71, REP2, miR159, miR160, miR160?, miR169, miR173) in total (left) and @HA immunoprecipitated (IP; right) fractions from SUC:SUL and 35S:P19HA/SUC:SUL rosette leaves was obtained by sequential rounds of probing and stripping the same membrane. The three IP samples (#1, #2, #3) correspond to three technical replicates obtained from a pool of ten 35S:P19HA/SUC:SUL plants. (B) Western analysis of P19HA accumulation in total (left) and @HA IP (right) fractions obtained from the plants described in (A). (C) Northern analysis of small RNAs in total (left) and peroxisomal (perox.; right) fractions obtained from 35S:P19HA/SUC:SUL and 35S:P19HASKL/SUC:SUL plants. The small RNA species detected are the same as in (A). The three peroxisome samples (#1, #2, #3) correspond to three biological replicates of 35S:P19HASKL/SUC:SUL plants. (D) Western analysis of plant peroxisomal marker hydroxypyruvate reductase (@HPR) and P19HA/P19HASKL (@HA) in total (left) and peroxisomal (right) fractions obtained from the plants described in (C). Note that more peroxisomal protein was loaded to detect P19HA/P19HASKL than to detect HPR. Northern analyses were performed on high-resolution gels. Accumulation of snU6 was used as RNA loading control, while coomassie staining was used as protein loading control. Figure source data can be found with the Supplementary information.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 29479364
Journal: Front Plant Sci
Figure Number: 4A
Published Date: 2018-02-27
First Author: Incarbone, M., Ritzenthaler, C., et al.
Impact Factor: 5.435
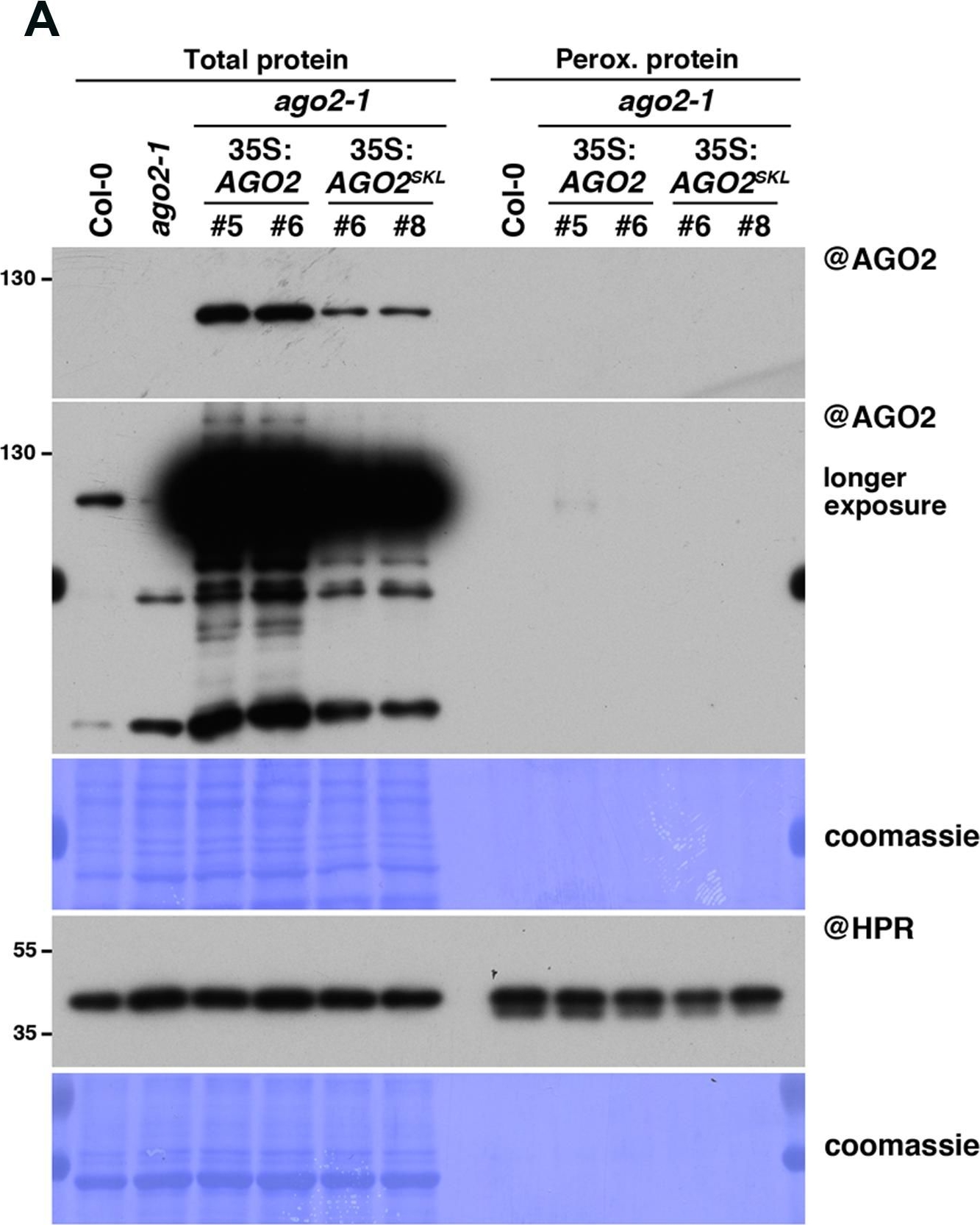
Open PublicationAGO2SKL is not imported into peroxisomes, while overexpression of AGO2 impacts the miR390/TAS3 pathway. (A) Western analysis of plant peroxisomal marker hydroxypyruvate reductase (@HPR) and AGO2/AGOSKL (@AGO2) in total (left) and peroxisome (right) fractions obtained from Col0, 35S:AGO2/ago2-1 (transgenic lines #5 and #6) and 35S:AGO2SKL/ago2-1 (transgenic lines #6 and #8) plants. Total proteins also include ago2-1. (B) Photos of the plants described in (A). (C) Northern analysis of small RNAs (miR159, miR173, miR390, miR390?, miR403, miR408, TAS1-derived siRNA (255), and TAS3-derived siRNA 5?D7(+) (TAS3)) in total leaf tissue of plants described in (A,B) was obtained by sequential rounds of probing and stripping the same membrane. Northern analysis was performed on a low-resolution gel. Accumulation of snU6 was used as RNA loading control, while coomassie staining was used as protein loading control. Figure source data can be found with the Supplementary Information.
- Background
-
Background: Hydroxypyruvate reductase (HPR) belongs to the D-isomer specific 2-hydroxyacid dehydrogenase family (oxidoreductases) and is involved in glycine, serine and theronine and glyoxylate and dicarboxylate metabolism. Synonymes: HPR, beta-hydroxypyruvate reductase, NADH:hydroxypyruvate reductase, D-glycerate dehydrogenase.
- Product Citations
-
Selected references: Bapatla et al. (2021). Modulation of Photorespiratory Enzymes by Oxidative and Photo-Oxidative Stress Induced by Menadione in Leaves of Pea (Pisum sativum). Plants 10, no. 5: 987. https://doi.org/10.3390/plants10050987
Korotaeva et al. (2018). Effect of Heat Hardening on Expression of Genes phb3 and phb4 and Accumulation of Phb Proteins in Green Leaves of Arabidopsis thaliana. Russian Journal of Plant Physiology, 65(5), 688-696, 2018 https://doi.org/10.1134/s1021443718040039
Farmer et al. (2013).Disrupting Autophagy Restores Peroxisome Function to an Arabidopsis lon2 Mutant and Reveals a Role for the LON2 Protease in Peroxisomal Matrix Protein Degradation. Plant Cell, Oct 31. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts - Reviews:
-
This product doesn't have any reviews.