1

Anti-GLDH | Galactono-1,4 lactone dehydrogenase
AS06 182 | Clonality: Polyclonal | Host: Rabbit | Reactivity: H. vulgare, G. max, A. sativa, Helianthus annuus, O. sativa, Z.mays
- Product Info
-
Immunogen: Recombinant C-terminal of Zea mays GLDH, UniProt: C0HFL3
Host: Rabbit Clonality: Polyclonal Purity: Total IgG. Protein G purified in PBS pH 7.4. Format: Lyophilized Quantity: 100 µl Reconstitution: For reconstitution add 100 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Western blot (WB) Recommended dilution: 1 : 5000 (WB) Expected | apparent MW: 68 kDa
- Reactivity
-
Confirmed reactivity: Avena sativa, Glycine max, Hordeum vulgare, Helianthus annuus, Oryza sativa, Zea mays Predicted reactivity: Arabidopsis thaliana, Zostera marina
Species of your interest not listed? Contact usNot reactive in: No confirmed exceptions from predicted reactivity are currently known - Application Examples
-
Application examples: 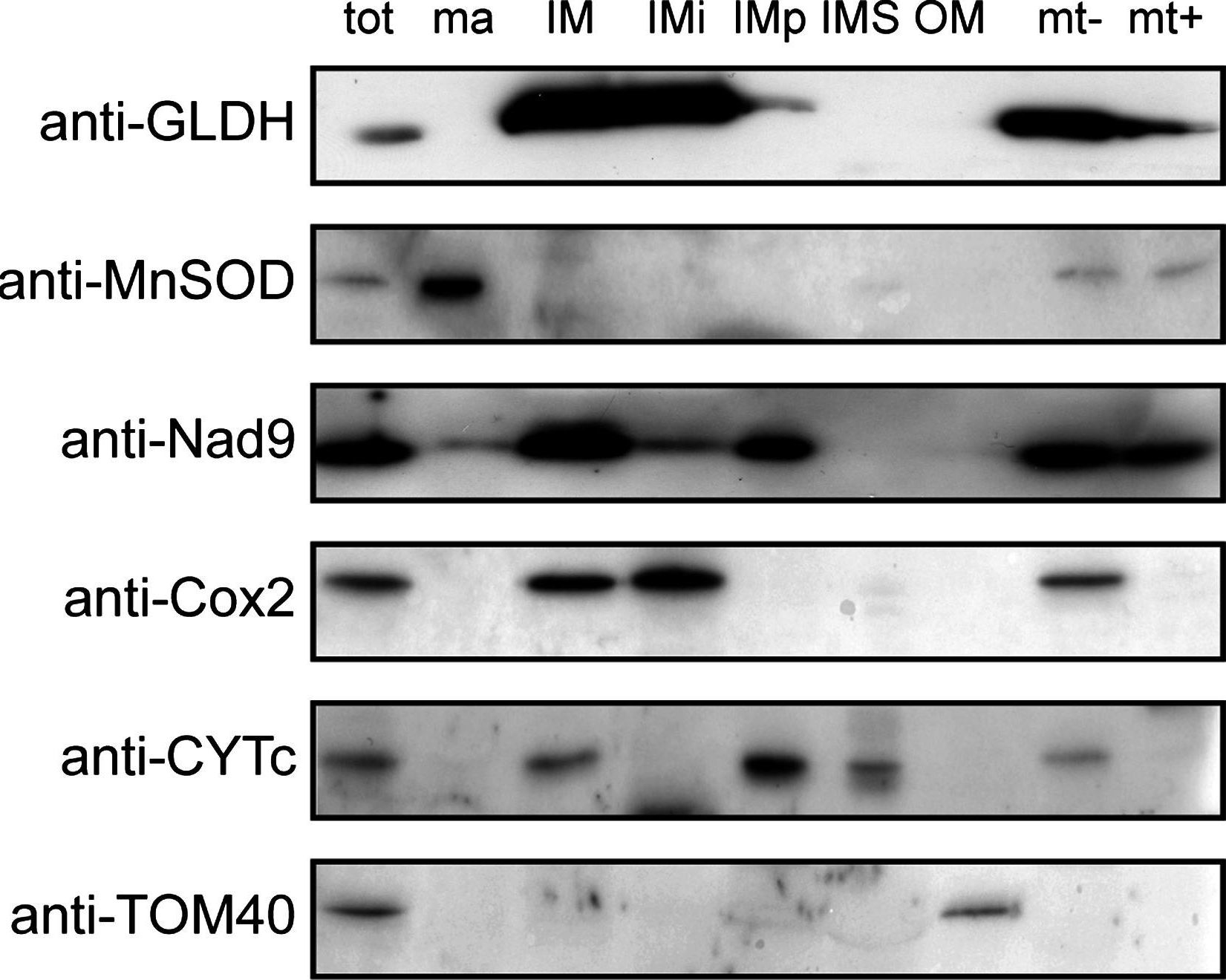
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 26520835
Journal: Plant Mol Biol
Figure Number: 1A
Published Date: 2016-01-01
First Author: Schimmeyer, J., Bock, R., et al.
Impact Factor: 3.808
Open PublicationSubmitochondrial localization of GLDH. Purified mitochondria (tot) were fractionated and the following submitochondrial compartments were isolated: matrix (ma), inner membrane (IM), intermembrane space (IMS) and outer membrane (OM). Integral proteins of IM (IMi) and peripheral proteins of IM (IMp) were separated by performing an alkali treatment of the IM (see “Material and methods”). In addition, mitochondria were swollen to obtain mitoplasts (swollen mitochondria lacking the outer membrane). Mitoplasts (mt?) were treated with proteinase K to digest the protein domains of the IM facing the IMS (mt+). The different fractions were separated by SDS-PAGE and western blots using anti-GLDH antibodies and antibodies against marker proteins for each compartment were performed
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 26520835
Journal: Plant Mol Biol
Figure Number: 2A
Published Date: 2016-01-01
First Author: Schimmeyer, J., Bock, R., et al.
Impact Factor: 3.808
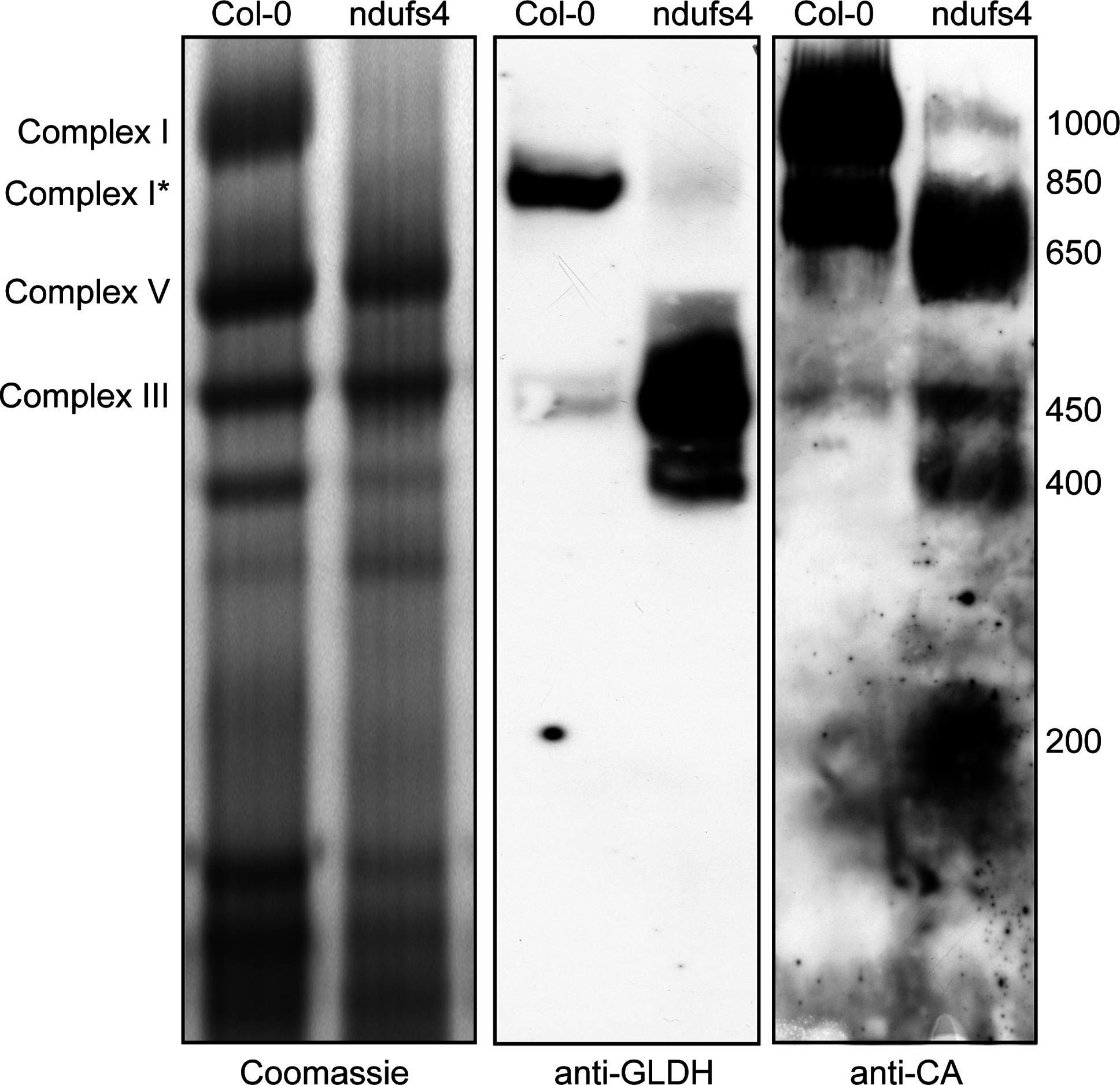
Open PublicationLocalisation of GLDH by BN-PAGE. Mitochondrial complexes of Col-0 and the complex I mutant ndufs4 were resolved by Blue-Native PAGE. Duplicate gels were run and either stained with Coomassie (left panel) or transferred onto a membrane for western blot analysis with anti-GLDH (middle panel) or anti-CA (right panel) antibodies. The position of selected respiratory complexes is indicated on the left. The size (in kDa) of complex I and the different assembly intermediate of complex I is indicated on the right
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 26520835
Journal: Plant Mol Biol
Figure Number: 4B
Published Date: 2016-01-01
First Author: Schimmeyer, J., Bock, R., et al.
Impact Factor: 3.808
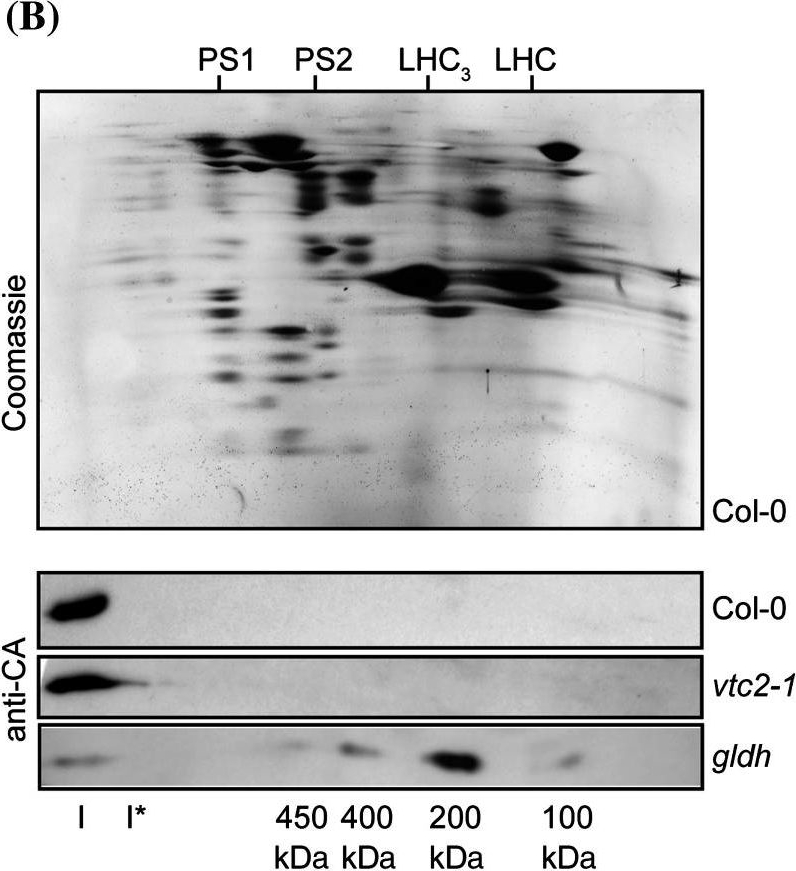
Open PublicationCharacterisation of complex I assembly in the gldh mutant. Total membranes were purified from Col-0 and the ascorbate-deficient mutants gldh and vtc2-1. a BN-PAGE analysis of membrane complexes. The left panel shows the Coomassie staining of the gel. The right panel shows the western blot analysis using anti-CA antibodies. The sizes of the marker bands of the molecular weight marker used for calibration of the gel are indicated in kDa on the left. The positions of complex I (I), complex I* (I*), photosystem 1 (PS1), photosystem 2 (PS2), the LHC trimers (LHC3) and LHC monomers (LHC) are indicated between both panels. b BN-SDS-PAGE analysis of membrane complexes. Top panel Coomassie staining of a representative gel (obtained for Col-0). The positions of photosystem 1 (PS1), photosystem 2 (PS2), the LHC trimers (LHC3) and LHC monomers (LHC) are indicated on the top. The bottom three panels show western blot analyses using anti-CA antibodies. The plant line analysed is indicated on the right. The positions of complex I (I) and the assembly intermediates detected in the gldh mutant are indicated in the bottom of the panel
- Additional Information
-
Additional information: Total IgG concentration is 6,8 μg/μl Additional information (application): Mitochondrial, membrane or meristematic fractions were shown to be richer in GLDH expression
- Background
-
Background: Galactono-1,4-lactone dehydrogenase (GLDH) is the enzyme which catalyses last step of ascorbic acid (AA) synthesis in the mitochondria of plant cells.
Alternative name: GaLDH - Product Citations
-
Selected references: Chen et al. (2019). Composition of Mitochondrial Complex I during the Critical Node of Seed Aging in Oryza sativa. Journal of Plant Physiology Volume 236, May 2019, Pages 7-14.
Schimmeyer et al. (2016). L-Galactono-1,4-lactone dehydrogenase is an assembly factor of the membrane arm of mitochondrial complex I in Arabidopsis. Plant Mol Biol. 2016 Jan;90(1-2):117-26. doi: 10.1007/s11103-015-0400-4. Epub 2015 Oct 31.
Ostaszewska-Bugajska et al. (2016). Changes in the OXPHOS system in leaf and root mitochondria of Arabidopsis thaliana subjected to long-term sulphur deficiency. Acta Physiologiae Plantarum 38:141.
Bartoli et al. (2005). Ascorbate content in wheat leaves is not determined by maximal L-galactono-1, 4-lactone dehydrogenase (GalLDH) activity under drought stress. Plant Cell Environ 28:1073-1081.
- Reviews:
-
This product doesn't have any reviews.
Accessories

AS04 052 | Clonality: Polyclonal | Host: Hen | Reactivity: A. thaliana (leaf extract and isolated mitochondria), B. oleracea, P. major, P. euryphylla, S. uniflora, S. dioica

AS05 074 | Clonality: Polyclonal | Host: Rabbit | Reactivity: A. thaliana,P.hybrida cv. Mitchell, P. grandiflora, S. oleracea, T. aestivum, V. faba | cellular [compartment marker] of mitochondrial matrix