1
Anti-ATG8 | Autophagy-related protein
AS14 2769 | Clonality: Polyclonal | Host: Rabbit | Reactivity: A. thaliana, A. madagascariensis, C. reinhardtii, Ch. dorsiventrale, G. hirsutum, H. lacustris, N. benthamiana, P. trichocarpa. S. lycopersicum, Z.mays
- Product Info
-
Immunogen: Fragment of recombinant ATG8 from Chlamydomonas reinhardtii, UniProt: A8JB85
Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Immunolocalization (IL), Western blot (WB) Recommended dilution: 1 : 1000 (IL), 1 : 1000-1 : 2000 (WB) Expected | apparent MW: 15.2 | 15 kDa - Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Aponogeton madagascariensis, Chlamydononas reinhardtii, Chlorococcum dorsiventrale, Gossypim hirsutum, Haematococcus lacustris, Nicotiana benthamiana, Populus trichocarpa, Solanum lycopersicum, Zea mays Predicted reactivity: Ananas comosus, Brassica napus, Hordeum vulgare, Micromonas sp., Nelumbo nucifera, Oryza sativa, Panicum hallii, Parachlorella kessleri, Phoenix dactylifera, Pyrus x bretschneideri, Physcomitrium patens, Pinus sitchensis, Solanum tuberosum, Triticum aestivum, Volvox carteri
Species of your interest not listed? Contact usNot reactive in: Cuscuta chinensis, Symbiodinium sp. - Application Examples
-
 Anti-CrATG8 antibodies detect 1 ng of recombinant CrATG8 protein.
Anti-CrATG8 antibodies detect 1 ng of recombinant CrATG8 protein.
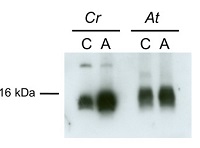
30 µg of total protein from Chlamydomonas reinhardtii , control (C), autophagy induced (A), extracted with lysis buffer according to Perez-Perez et al. 2010 (Plant Physiology 152: 1874-1888) were separated on 15 % SDS-PAGE and blotted 1h to nitrocellulose membrane using semi-dry or tank transfer. Blots were blocked with 5% milk for 1h at room temperature (RT) with agitation. Blot was incubated in the primary antibody at a dilution of 1:1000 for 1h at RT with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at RT with agitation. Blot was incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated, from Agrisera AS09 602, diluted to 1:25 000) for 1h at RT with agitation. The blot was washed as above and developed for 5 min with chemiluminescent detection reagent, according to the manufacturer's instructions. Exposure time was 45 seconds.Courtesy of Dr. María Esther Pérez-Perez, IBVF, Spain
15 µg of total protein from Chlamydomonas reinhardtii and Arabidopsis thaliana were separated on 15 % SDS-PAGE and blotted 1h to nitrocellulose membrane using semi-dry transfer. Blots were blocked with 5 % dry milk in PBS for 1h at room temperature (RT) with agitation. Blot was incubated in the primary antibody at a dilution of 1:1 000 over night at 4 ºC with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at RT with agitation. Blot was incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated, AS09 602 from Agrisera) diluted to 1:10 000 in 5 % dry milk for 1h at RT with agitation. The blot was washed as above and developed for 5 min with chemiluminescent detection reagent, according to the manufacturer's instructions. Exposure time was 60 seconds.Courtesy of Dr. María Esther Pérez-Pérez and Ana M. Laureano-Marín, IBVF, SpainApplication examples: 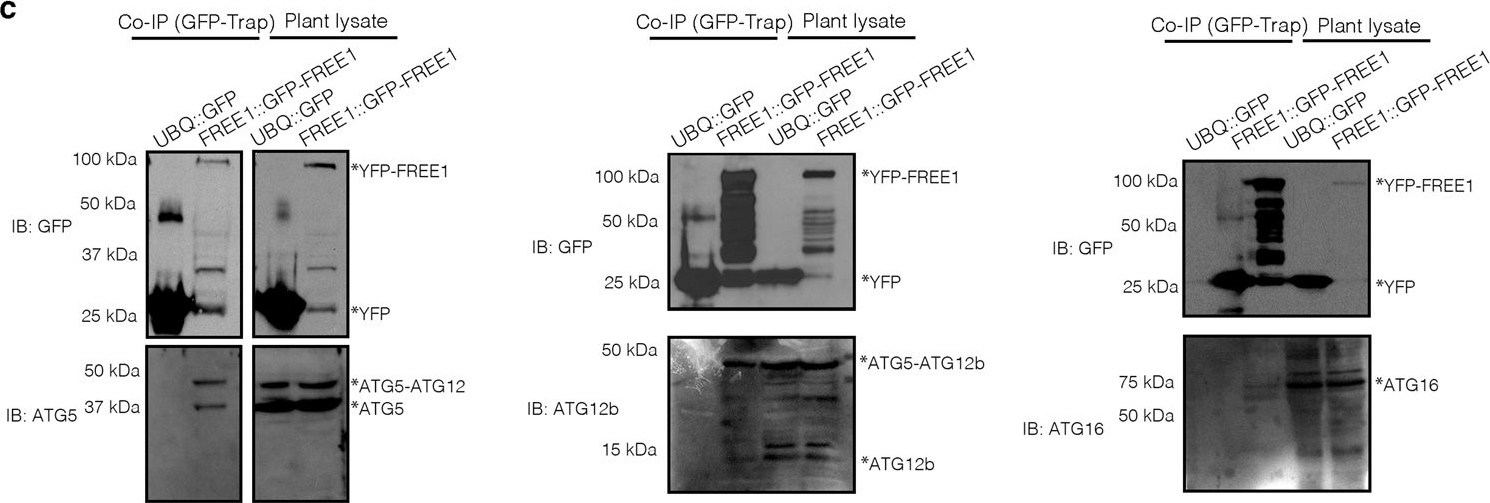
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 36997511
Journal: Nat Commun
Figure Number: 2C
Published Date: 2023-03-30
First Author: Zeng, Y.
Impact Factor: 15.405
Open PublicationFREE1 interacts with ATG conjugation system and ESCRT-III under starvation conditions.a Transgenic Arabidopsis seedlings expressing YFP-FREE1 upon autophagy induction by carbon starvation for at least 18 h were subjected to GFP-Trap assay for IP-MS analysis. Identified FREE1-interacting partners were analyzed using STRING. b Immunofluorescence labeling of the endogenous ATG conjugation system proteins ATG5, ATG12b or ATG16 in Arabidopsis protoplasts transfected with GFP-FREE1 for 24 h before confocal laser scanning microscopy (CLSM) observation. White dash boxes indicate the 5 X enlarged areas. Scale bars, 10 μm. c GFP-Trap and co-IP analysis of GFP-FREE1 with endogenous ATG5, ATG12b, or ATG16 using native promoter driven GFP-FREE1 transgenic plants. Arabidopsis transgenic plants expressing GFP or GFP-FREE1 driven by the native promoter were subjected to protein extraction and IP with GFP-Trap, followed by western blot analysis with indicated antibodies. d Structural TEM analysis of high-pressure freezing/frozen substituted atg5-1 mutant plant root tips upon BTH treatment for 8 h. Black dash boxes indicated the 5 X zoom-in areas. Arrowheads indicated examples of the unsealed autophagosomal structures. Scale bars, 500 nm. e 5 X zoom-in areas from d. Arrowheads indicated examples of the unsealed autophagosomal structures. Scale bars, 500 nm. f Quantification analysis of the ratio of the unclosed autophagosomal structures (APs) in Col-0 and atg5-1 mutants per cross-cell section. Means±SD; n = 10 (Col-0) and n = 17 (atg5-1) cells, two-tailed unpaired t test; ***p < 0.001. All the imaging analysis and immunoblots were repeated at least for three times with similar results.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 36997511
Journal: Nat Commun
Figure Number: 4J
Published Date: 2023-03-30
First Author: Zeng, Y.
Impact Factor: 15.405
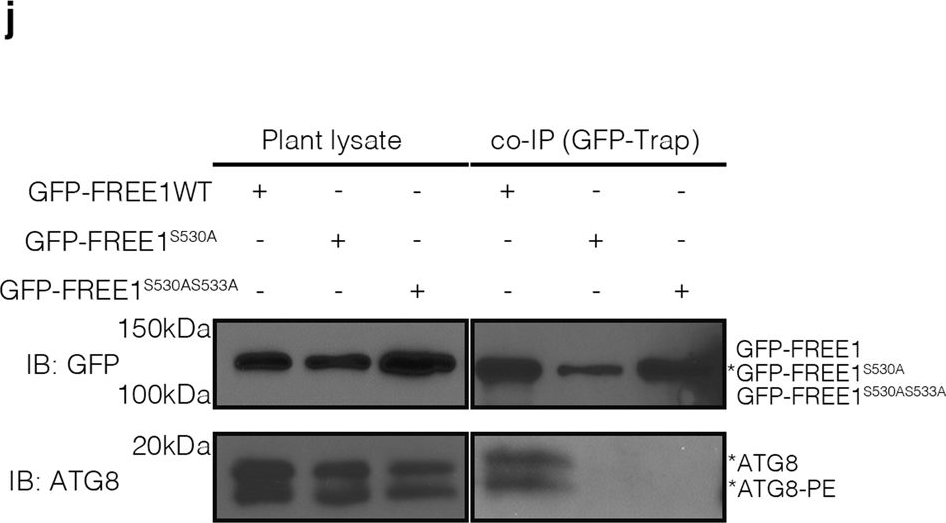
Open PublicationKIN10 phosphorylates FREE1 to orchestrate ESCRT machinery function in autophagosome closure.a Transient expression and confocal analysis of the spatial relationship between FREE1 and KIN10 in Arabidopsis leaf protoplasts. The transfected leaf protoplasts were cultured for 24 h before CLSM observation. Scale bars, 10 μm. b FRET analysis of CFP-FREE1 with KIN10-YFP. Arabidopsis protoplasts co-expressing CFP and YFP (negative control), CFP-YFP (positive control), or CFP-FREE1 and KIN10-YFP were cultured for 24hr before FRET analysis. Means±SD; n = 16 (CFP/YFP), n = 11 (CFP-YFP), and n = 16 (CFP-FREE1/KIN10-YFP) individual puncta per experimental group, one-way analysis of variance (ANOVA), followed by Tukey’s multiple test; ***p < 0.001. The middle lines of the boxes represent the medians of datasets. The upper and bottom lines of the boxes are respectively the upper quantile and the lower quantile of the data. The whiskers mark the upper and lower limits of these datasets, respectively. c Y2H analysis between AD-KIN10 and BD-FREE1231–601 (the truncated FREE1 without self-activation on BD vector). d GFP-Trap and co-immunoprecipitation (co-IP) analysis of KIN10-6HA and YFP-FREE1 in Arabidopsis protoplasts. Arabidopsis protoplasts co-expressing KIN10-6HA with YFP-FREE1, or YFP only were subjected to protein extraction and IP with GFP-Trap, followed by immunoblotting with indicated antibodies. e GFP-Trap and co-IP of KIN10 and FREE1 using transgenic plants expressing KIN10-GFP under nitrogen starvation (-N) for at least 18hrs, followed by immunoblotting using GFP and FREE1 antibodies. f Quantification analysis of the ratio of co-immunoprecipitated FREE1 by KIN10 shown in e. Means±SD; n = 4 individual experiments, two-tailed unpaired t test, ***p < 0.001. g In vivo phosphorylation of FREE1 upon nitrogen starvation. GFP-FREE1/free1 transgenic plants were subjected to mock or nitrogen starvation for at least 18 h, followed by protein extraction and IP with GFP-Trap, and subsequent immunoblotting detection by Phos-tag biotin and the indicated antibodies. h Semi-in vitro phosphorylation assay of FREE1, FREE1S530A and FREE1S530AS533A proteins by KIN10-GFP purified from the transgenic plants expressing KIN10-GFP. KIN10 was enriched from transgenic seedlings expressing KIN10-GFP by immunoprecipitation with anti-GFP and then used to phosphorylate FREE1, FREE1S530A or FREE1S530AS533A tagged with His-sumo, which was further detected by Phos-tag biotin. i Semi-in vitro phosphorylation assay of FREE1, FREE1S530A and FREE1S530AS533A proteins by KIN10-GFP purified from the transgenic plants expressing KIN10-GFP in snrk2.2/2.3 mutants. KIN10 was enriched from transgenic seedlings expressing KIN10-GFP in snrk2.2/2.3 mutants background by immunoprecipitation with anti-GFP and then used to phosphorylate FREE1 or FREE1S530A or FREE1S530AS533A tagged with His-sumo, which was further detected by Phos-tag biotin. j In vivo interaction between ATG8 and FREE1, FREE1S530A, or FREE1S530AS533A mutant variants. GFP-FREE1, GFP-FREE1S530A, and GFP-FREE1S530AS533A transgenic plants were subjected to nitrogen starvation for at least 18 h, followed by protein extraction and IP with GFP-Trap, and subsequent immunoblotting with indicated antibodies. k Transient co-expression of GFP-FREE1 or GFP-FREE1S530A with mCherry-ATG8i in Arabidopsis protoplasts. The transfected protoplasts were cultured for 24 h before CLSM observation. Scale bars, 10 μm. All the imaging analysis and immunoblots were repeated at least for three times with similar results.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 36997511
Journal: Nat Commun
Figure Number: 3E
Published Date: 2023-03-30
First Author: Zeng, Y.
Impact Factor: 15.405
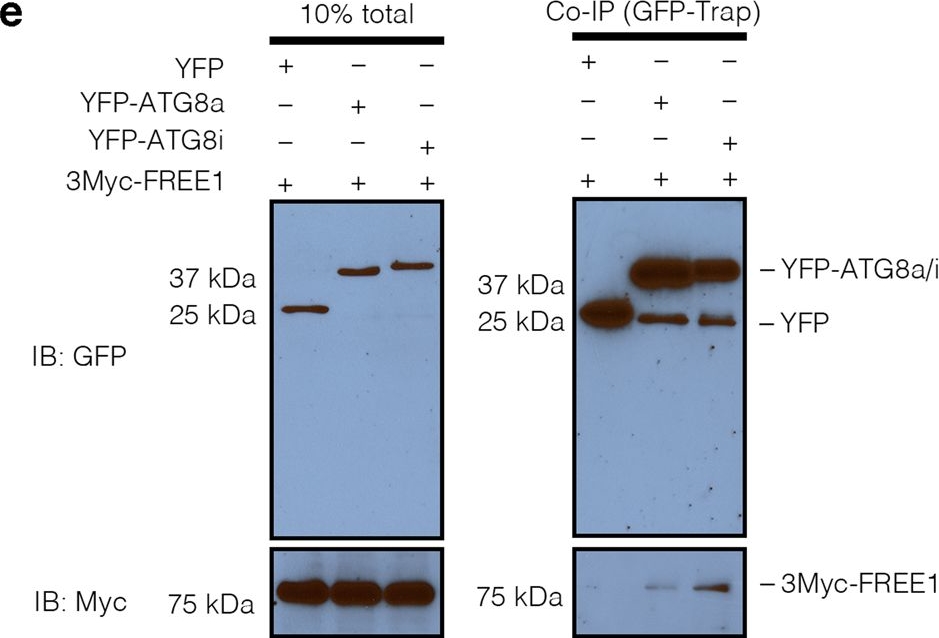
Open PublicationFREE1 specifically interacts with the distinct paralogs of ATG8.a Y2H analysis of BD-FREE1231–601 (the truncated FREE1 without self-activation on BD vector) with AD-ATG8 isoforms. b Evolutionary relationships of ATG8 isoforms. The optimal tree with the sum of branch length=4.32642842 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. c Transient expression of CFP-FREE1 with specific ATG8 isoforms YFP-ATG8a and YFP-ATG8i in Arabidopsis protoplasts, the transfected protoplasts were cultured for 24 h before CLSM observation. White dash boxes indicate the 2.5 X enlarged insets. Scale bars, 10 μm. dArabidopsis protoplasts transfected with FREE1, ATG5 and ATG8a or ATG8i were subjected to structure illuminated microscopy (SIM) analysis. Scale bars, 1 μm. e GFP-Trap and co-immunoprecipitation (co-IP) analysis of 3Myc-FREE1 with YFP-ATG8a or YFP-ATG8i using Arabidopsis protoplasts. Arabidopsis protoplasts co-expressing 3Myc-FREE1 with YFP-ATG8a, YFP-ATG8i, or YFP only were subjected to protein extraction and co-IP with GFP-Trap, followed by immunoblotting with indicated antibodies. f FRET analysis between CFP-FREE1 and YFP-ATG8a or YFP-ATG8i in Arabidopsis protoplasts. Arabidopsis protoplasts were transfected with CFP and YFP (negative control), CFP-YFP (positive control), CFP-FREE1 and YFP-ATG8a, or CFP-FREE1 and YFP-ATG8i, followed by cultured for 24 h before FRET analysis. Means±SD; n = 14 (CFP/YFP), n = 14 (CFP-YFP), n = 28 (CFP-FREE1/YFP-ATG8a), and n = 33 (CFP-FREE1/YFP-ATG8i) individual puncta per experimental group, one-way analysis of variance (ANOVA), followed by Dunnett’s multiple comparisons test; *p < 0.05; **p < 0.01. The middle lines of the boxes represent the medians of datasets. The upper and bottom lines of the boxes are the upper quantile and the lower quantile of the data, respectively. The whiskers mark the upper and lower limits of these datasets, respectively. All the imaging analysis and immunoblots were repeated at least for three times with similar results.
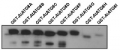
Reactant: Chlamydomonas reinhardtii (Green Alga)
Application: Western Blotting
Pudmed ID: 32765574
Journal: Front Plant Sci
Figure Number: 6B
Published Date: 2020-08-09
First Author: Chen, X. D., Liu, Y., et al.
Impact Factor: 5.435
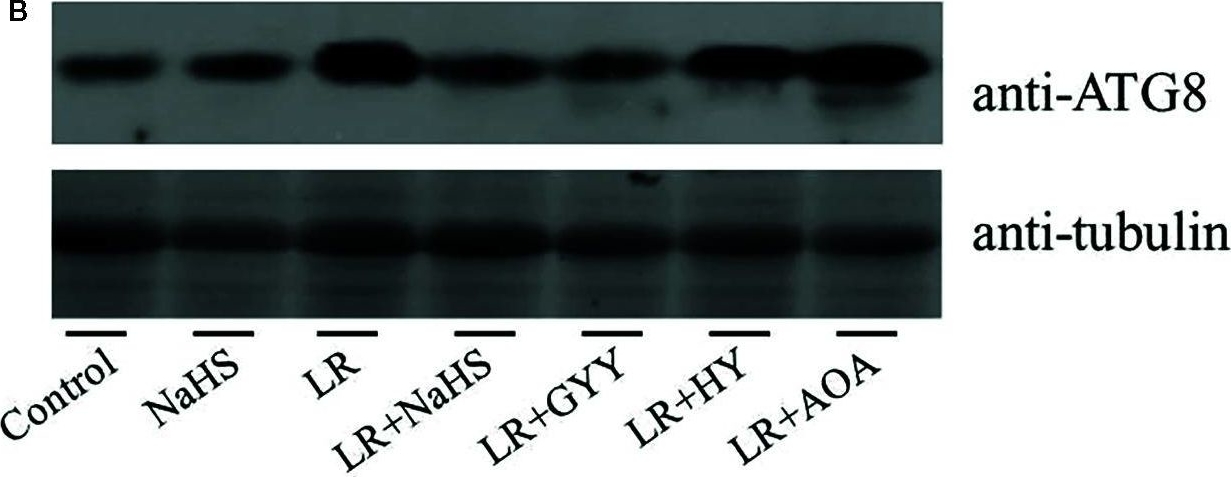
Open PublicationEffects of different H2S donors or H2S biosynthesis inhibitors on cell autophagy capability of C. reinhardtii under MC-LR treatment. (A) Effects of H2S biosynthesis scavenger or inhibitor on MC-LR-induced expression of ATG1, AGT7, AGT8, and ATG101. Here, 3-d-old C. reinhardtii cells were treated with 100 nM MC-LR with or without H2S artificial donor NaHS (100??M), GYY4137 (GYY, 10??M), H2S scavenger hypotaurine (HY, 10??M), or H2S biosynthesis enzyme inhibitor aminooxyacetic acid (AOA, 1?mM) for 12 h, with transcriptional levels of ATG1, AGT7, AGT8, and ATG101 then measured by qRT-PCR analysis. Values are means ± SD of three biological replicates. (B) Effects of H2S biosynthesis scavenger or inhibitor on MC-LR-induced protein accumulation of ATG8. Here, 3-d-old C. reinhardtii cells were treated with 100 nM MC-LR with or without H2S artificial donor for 3 d, with protein accumulation of ATG8 measured by Western blotting using anti-ATG8 antibody. Anti-Tubulin was used as the loading control.
- Additional Information
-
Additional information: This product can be sold containing ProClin if requested.
This antibody does not recognize all isoforms into the same degree.
This antibody is recognizing 1 ng of recombinant CrATG8.
Antigen used to elicit this antibody is conserved from 70-80% in following ATG protein from Arabidopsis thaliana: ATG8a UniProt: Q8LEM4 ATG8B UniProt: Q9XEB5 ATG8c UniProt: Q8S927 ATG8d UniProt: Q9SL04, ATG8e UniProt: Q8S926 ATG8f UniProt: Q8VYK7 and conserved below 70% in: ATG8g UniProt: Q9LZZ9 ATG8h Uniprot: Q8S92Additional information (application): For Arabidopsis thaliana the signal obtained using ATG8 antibodies is cleaner in case of roots compare to leaf material. For best results please follow extraction protocol described in Álvarez et al. (2012). ATG8 signal corresponds to the two bands of 17 kDa.Preparation of a cell extract from Arabidopsis thaliana:A. Plants were first subjected to autophagy activating conditions: nutrient (nitrogen or carbon) limitation or oxidative stress in order to activate this degradative process.B. Total protein extracts can be obtained as described by Álvarez. Leaves are grinded in liquid nitrogen with a minimal volume of extraction buffer (100 mM Tris-HCl pH 8, 400 mM sucrose, 1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mg/ml sodium deoxycholate, 10 µg/ml of leupeptin, 10 µg/ml of pepstatin A, 4% (v/v) protease inhibitor cocktail from Roche).C. Cell debris is removed by centrifuging at 500 g for 10 min at 4°C.
Important note:It is recommendable to use bigger gels in order to get a better resolution of ATG8 bands. Midi-protean gels are better than mini-gels. There are 9 ATG8 isoforms and this antibody will likely recognizes all of them.
For immunolocalization protocol, please inquire. - Background
-
Background: ATG8 (Autophagy-related protein 8) is involved in degradation and recycling of intracellular components in a process of autophagy. ATG8 is a molecular autophagy marker in Chlamydomonas reinhardtii (Pérez-Pérez et al. 2010, Plant Physiol. 152: 1874-88). - Product Citations
-
Selected references: Zheng et al. (2025). ATG8ylation-mediated tonoplast invagination mitigates vacuole damage. Nat Commun . 2025 Jul 18;16(1):6621. doi: 10.1038/s41467-025-62084-3.
Supriya et al. (2025). Melatonin Bridges Timekeeping and Survival: Coordinating Diurnal Rhythms, Autophagy, and Energy Balance in Upland Cotton. Plant Stress, 100969.
Li et al. (2024). Stress granules sequester autophagy proteins to facilitate plant recovery from heat stress. Nat Commun . 2024 Dec 30;15(1):10910. doi: 10.1038/s41467-024-55292-w.
Li et al. (2024). Gamma-aminobutyric acid as a regulator of astaxanthin production in Haematococcus lacustris under salinity: Exploring physiology, signaling, autophagy, and multi-omics landscape. Bioresour Technol. 2024 Dec:413:131466.doi: 10.1016/j.biortech.2024.131466. Epub 2024 Sep 12.
Michalak et al. (2024). Conserved autophagy and diverse cell wall composition: Unifying features of vascular tissues in evolutionarily distinct plants.Ann Bot. 2024 Apr 23;133(4):559-572. doi: 10.1093/aob/mcae015.
Lan et al. (2024).Clathrin Light Chains negatively regulate plant immunity by hijacking the autophagy pathway. Plant Commun. 2024 Apr 30:100937.doi: 10.1016/j.xplc.2024.100937.
Testi et al. (2024). The Phytophthora parasitica effector AVH195 interacts with ATG8, attenuates host autophagy, and promotes biotrophic infection. BMC Biol. 2024 Apr 29;22(1):100.
Michalak et al. (2024). Conserved autophagy and diverse cell wall composition: Unifying features of vascular tissues in evolutionarily distinct plants. Ann Bot. 2024 Feb 7:mcae015. (immunofluorescence) doi: 10.1093/aob/mcae015.
Llamas et al. (2023). In planta expression of human polyQ-expanded huntingtin fragment reveals mechanisms to prevent disease-related protein aggregation. Nat Aging. 2023 Nov;3(11):1345-1357.doi: 10.1038/s43587-023-00502-1.
Miklaszewska et al. (2023).CALEOSIN 1 interaction with AUTOPHAGY-RELATED PROTEIN 8 facilitates lipid droplet microautophagy in seedlings.Plant Physiol. 2023 Nov 22;193(4):2361-2380.doi: 10.1093/plphys/kiad471.
Lee et al. (2023).Three consecutive cytosolic glycolysis enzymes modulate autophagic flux. Plant Physiol. 2023 Oct 26;193(3):1797-1815. doi: 10.1093/plphys/kiad439.
Zhao et al (2023) A non-canonical role of ATG8 in Golgi recovery from heat stress in plants. Nat Plants. 2023 May;9(5):749-765. doi: 10.1038/s41477-023-01398-w.
Zeng et al. (2023) The plant unique ESCRT component FREE1 regulates autophagosome closure. Nat Commun. 2023 Mar 30;14(1):1768. doi: 10.1038/s41467-023-37185-6.
Sun et al. (2022) Genome of Paspalum vaginatum and the role of trehalose mediated autophagy in increasing maize biomass. Nat Commun. 2022;13(1):7731. Published 2022 Dec 13. doi:10.1038/s41467-022-35507-8
Cao et al. (2022) Autophagic pathway contributes to low-nitrogen tolerance by optimizing nitrogen uptake and utilization in tomato. Hortic Res. 2022 Mar 23;9:uhac068. doi: 10.1093/hr/uhac068. PMID: 35669705; PMCID: PMC9164271.
Samperna et al (2022). Cyclopaldic Acid, the Main Phytotoxic Metabolite of Diplodia cupressi, Induces Programmed Cell Death and Autophagy in Arabidopsis thaliana. Toxins (Basel). 2022 Jul 11;14(7):474. doi: 10.3390/toxins14070474. PMID: 35878212; PMCID: PMC9325063.
Zharova et al. (2022) Role of Autophagy in Haematococcus lacustris Cell Growth under Salinity. Plants. 2022; 11(2):197. https://doi.org/10.3390/plants11020197 (immunofluorescence)
Mishra et al. (2021) Interplay between abiotic (drought) and biotic (virus) stresses in tomato plants. Mol Plant Pathol. 2021 Dec 30. doi: 10.1111/mpp.13172. Epub ahead of print. PMID: 34970822.
De Brasi-Velasco et al. (2021). Autophagy Is Involved in the Viability of Overexpressing Thioredoxin o1 Tobacco BY-2 Cells under Oxidative Conditions. Antioxidants. 2021; 10(12):1884.
Sun et al. (2021) Mechanistic insights into an atypical interaction between ATG8 and SH3P2 in Arabidopsis thaliana. Autophagy. 2021 Oct 17:1-17. doi: 10.1080/15548627.2021.1976965. Epub ahead of print. PMID: 34657568.
Li et al. (2021) The unfolded protein response plays dual roles in rice stripe virus infection through fine-tuning the movement protein accumulation. PLoS Pathog. 2021 Mar 4;17(3):e1009370. doi: 10.1371/journal.ppat.1009370. Epub ahead of print. PMID: 33662041.
Li et al. (2021) Chloroplast proteotoxic stress-induced autophagy is involved in the degradation of chloroplast proteins in Chlamydomonas reinhardtii. Plant Cell Physiol. 2021 Feb 17:pcab029. doi: 10.1093/pcp/pcab029. Epub ahead of print. PMID: 33594417.
Kazibwe et al. (2020). TOR mediates the autophagy response to altered nucleotide homeostasis in a ribonuclease mutant. J Exp Bot. 2020 Sep 9;eraa410.doi: 10.1093/jxb/eraa410.
Upadhyaya and Jagadeeshwar Rao (2019). Reciprocal regulation of photosynthesis and mitochondrial respiration by TOR kinase in Chlamydomonas reinhardtii. Plant Direct Volume 3, Issue 11.
Shull et al. (2019). Anatase TiO2 nanoparticles induce autophagy and chloroplast degradation in thale cress (Arabidopsis thaliana). Environ Sci Technol. 2019 Jul 29. doi: 10.1021/acs.est.9b01648.
Wojciechowska et al. (2018). Autophagy counteracts instantaneous cell death during seasonal senescence of the fine roots and leaves in Populus trichocarpa. BMC Plant Biol. 2018 Oct 29;18(1):260. doi: 10.1186/s12870-018-1439-6. (immunolocalization)
Chen et al. (2016). The role of nitric oxide signalling in response to salt stress in Chlamydomonas reinhardtii. Planta. 2016 Sep;244(3):651-69. doi: 10.1007/s00425-016-2528-0. Epub 2016 Apr 26.
Gorovits et al. (2016). Tomato yellow leaf curl virus confronts host degradation by sheltering in small/midsized protein aggregates. Virus Res. 2016 Feb 2;213:304-13. doi: 10.1016/j.virusres.2015.11.020. Epub 2015 Dec 1.
- Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
Oxygenic photosynthesis poster by prof. Govindjee and Dr. Shevela
Z-scheme of photosynthetic electron transport by prof. Govindjee and Dr. Björn and Dr. Shevela - Reviews:
-
Marie Bertrand | 2023-07-26We used this antibody on chlamydomaonas. With a 1:2000 dilution we get a verry good signal. I recommand it !Xiaohong Zhuang | 2022-11-28It works quite well with our samples. The signal is quite strong and specific as described.Zeng Yonglun | 2021-12-06Nice antibody with relatively high specificity with lower exposure times.Peter Ma | 2020-08-12This antibody works very well for our samples. The membrane proteins from the Arabidopsis roots are subjected to 15% SDS-PAGE with 6M urea and then detected with this antibody (1:5000). The results shows that this antibody is quite specific and sensitive.Natalia | 2018-11-13Clean signal in WB as well as during localization in confocal microscopyAdrian Dauphinee | 2016-11-16Worked well in Aponogeton madagascariensis leaf tissues for immunolocalization experiments and western blotting.Dr. Rena Gorovits | 2015-04-14Obvious clean signal in proteins, extracted from stressed tomatoes.