1


Anti-UGPase | UDP-glucose pyrophosphorylase (cytoplasm marker)
AS05 086 | Clonality: Polyclonal | Host: Rabbit | Reactivity: A. thaliana, C. annuum, C. sativus, F. margarita Swingle, F. arundinacea, H. vulgare, L. esculentum, L. chilense, Malus x domestica Borkh. c.v. Fuji, M. polymorpha, Medicago truncatula, N. tabacum, O. sativa, P. glauca, Populus sp., S. tuberosum, S. sogarandinum, Triticum | cellular [compartment marker] of cytoplasm
Replaced by AS14 2813
- Product Info
-
Immunogen: Recombinant overexpressed and purified from E.coli UGPase Arabidopsis thaliana UGPase, UniProt: A0A1I9LT02
TAIR: AT3G03250Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Immunolocalization (IL), Western blot (WB) Recommended dilution: 1 : 1500 (IL), 1 : 1000-1 : 5000 (WB) Expected | apparent MW: 51.6 kDa - Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Brassica rapa, Capsicum annuum, Fortunella margarita Swingle, Citrus sinensis, Cucumis sativus, Festuca arundinacea, Hordeum vulgare, Lycopersicum esculentum, Lycopersicum chilense, Malus x domestica Borkh. c.v. Fuji,, Marchantia polymorpha, Medicago truncatula, Mesemryanthenum crystallinum, M.vaginalis, Nicotiana benthamiana, Nicotiana tabacum, Oryza sativa, Phaseolus vulgaris, Picea glauca, Populus sp., Solanum lycopersicum, Solanum tuberosum, Solanum sogarandinu, Triticum aestivu, Zea mays Predicted reactivity: Amorpha fruticosa, Bambusa oldhamii, Brachypodium distachyon, Brassica pekinensis, Capsella rubella, Cucumis melo, Eucalyptus grandis, Glycine max, Glycine soja, Gossipium hirsutum, Jatropha curcas, Pinus taeda, Populus tremula, Ricinus communis, Saccharum officinarum, Sorghum bicolor, Theobroma cacao, Zea mays, Vitis vinifera
Species of your interest not listed? Contact usNot reactive in: C. merolae, diatoms
- Application Examples
-
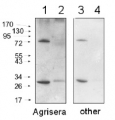
UGPase was used as a negative control for cytoplasm contamination in chloroplast fraction.
Total protein (first 2 lanes to the left) or chloroplast protein (2 lanes to the right) of Arabidopsis thaliana were extracted with 100 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1 mM DTT, 1 mM PMSF, protease inhibitor cocktail, denatured with Laemmli buffer (62.5 mM Tris–HCl pH 6.8, 2% SDS, 10% glycerol, 100 mM DTT, 0.1% β-mercaptoethanol) at 95°C 5 min and separated on 8% SDS-PAGE and blotted 1h to PVDF, using wet transfer. Blot was blocked with 5% milk for 1h/RT or 4°C/ON with agitation. Blot was incubated in the primary antibody at a dilution of 1: 2 000 for 1h/RT with agitation in TBS-T. The antibody solution was decanted and the blot was washed 3 times for 10 min in TBS-T at RT with agitation. Blot was incubated in Calbiochem matching secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated) diluted to 1:25 000 in for 1h/RT with agitation. The blot was washed as above and developed for 10 min with Clarity Western ECL Substrate (Biorad) and visualised with chemiluminescence CCD camera Fluorchem SP (Gel Biosciences). Exposure time was 1-5 minutes.
This antibody is required to be used on chloroplast fraction. There is no signal in the total cell extract.
Courtesty of Dr. Aleksandra Kwaśnik, Warsaw University, Poland
.jpg)
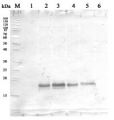
A 1-year-old greehouse grown plant was dissected into different tissues, which were then used for enzyme assays and immunoblot analyses. Equal amounts of total protein (7.5 μg) were loaded on each lane. SDS-PAGE was run on a 7.5% gel. Immunoblot was done using PVDF transfer membrane. Primary antibodies against barley UGPase were used in 1: 1000 dilution. Secondary antibodies (Rabbit IgG, HRP conjugated) were used at 1:10 000.
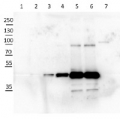
ff - female flower, mf - male flower, yl - young leaf, ml - mature leaf, sbk - stem bark, sph - stem phloem and cambium, sxy - stem xylem, rxy - root xylem

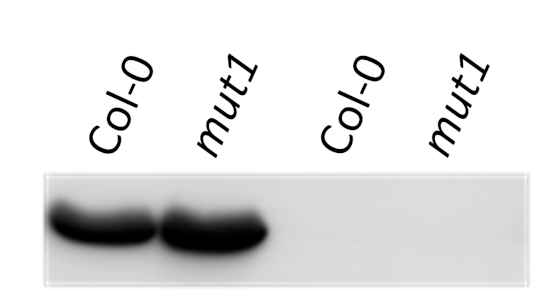
15 µg of total soluble protein extract from leaves and stems of Solanum tuberosum (1), Solanum sogarandinum (2), Lycopersicum esculentum (3), Lycopersicum chilense (4) , Arabidopsis thaliana (5) , Cucumis sativus (6) , Festuca arundinacea (7) , Nicotiana tabacum (8) and Capsicum annuum (9) were separated on 10% SDS-PAGE and blotted onto nitrocellulose . After blocking with 5% milk in TBST , blots were incubated with the primary antibody at a dilution of 1:1500 in TBST for 1h at room temperature. Following incubation and wash steps, blots were incubated with secondary Anti-Rabbit IgG , Alkaline Phosphatase Conjugate for 1 hour at a dilution of 1:40000 . Blots were developed with the alkaline phosphatase detection system using NBT/BCIP.
Courtesy of Bartosz Szabala, Institute of Plant Genetics, Polish Academy of Science .
Application examples: 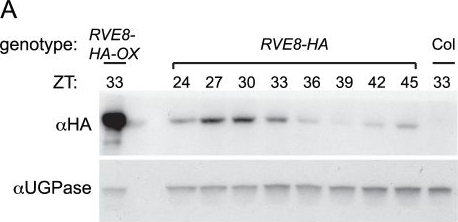
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 21483796
Journal: PLoS Genet
Figure Number: 4A
Published Date: 2011-03-01
First Author: Rawat, R., Takahashi, N., et al.
Impact Factor: 5.334
Open PublicationRVE8 protein accumulates in the subjective afternoon.Col, RVE8::RVE8-HA and 35S::RVE8-HA seedlings were entrained in 12 hr light:12 hr dark for 6 days before being released to constant white light (55 mol m−2 s−1) (A) Plants were harvested at the indicated times and extracts were subjected to immunoblot analysis using either an anti-HA antibody (upper panel) or an anti-UGPase antibody (lower panel). (B) Data shown in panel (A) was quantified using ImageQuant software. Data are representative of two independent experiments.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 21689410
Journal: BMC Plant Biol
Figure Number: 8A
Published Date: 2011-06-20
First Author: Balsemão-Pires, E., Jaillais, Y., et al.
Impact Factor: 4.142
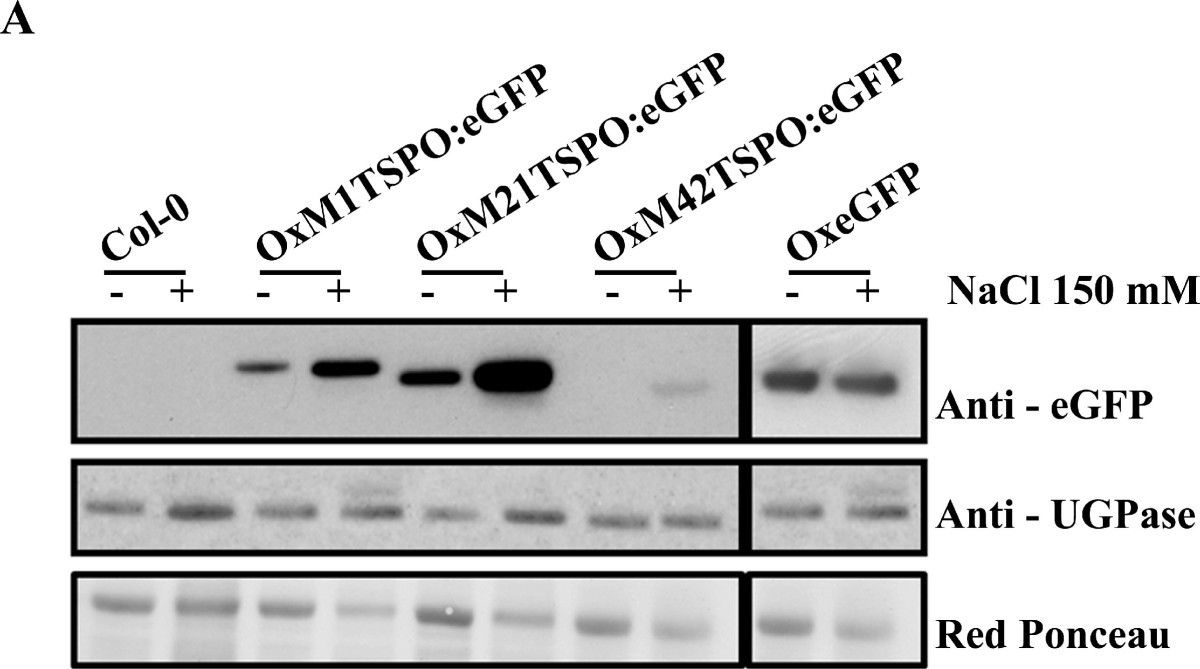
Open PublicationAtTSPO accumulation and chloroplast localization upon salt stress. (A) Immunoblot analysis of OxAtTSPO:eGFP (OxM1TSPO:eGFP, OxM21TSPO:eGFP and OxM42TSPO:eGFP) fusion proteins detected in plants with an antibody to GFP. Plants were untreated, or treated with 150 mM NaCl. As a control wild-type plants and plants over-expressing GFP (OxeGFP) seedlings were used. (B) Anti-GFP immunoblot of trypsinized chloroplasts from Arabidopsis plants either untreated or treated with 150 mM NaCl. Control immunoblots were probed with antibodies chloroplast proteins RuBisCo and D1; and to cytosolic UGPase. Each lane represents equal amounts of chloroplasts.
Reactant: Solanum lycopersicum (Tomato)
Application: Western Blotting
Pudmed ID: 28394025
Journal: New Phytol
Figure Number: 1B
Published Date: 2017-07-01
First Author: Howden, A. J. M., Stam, R., et al.
Impact Factor: 7.96
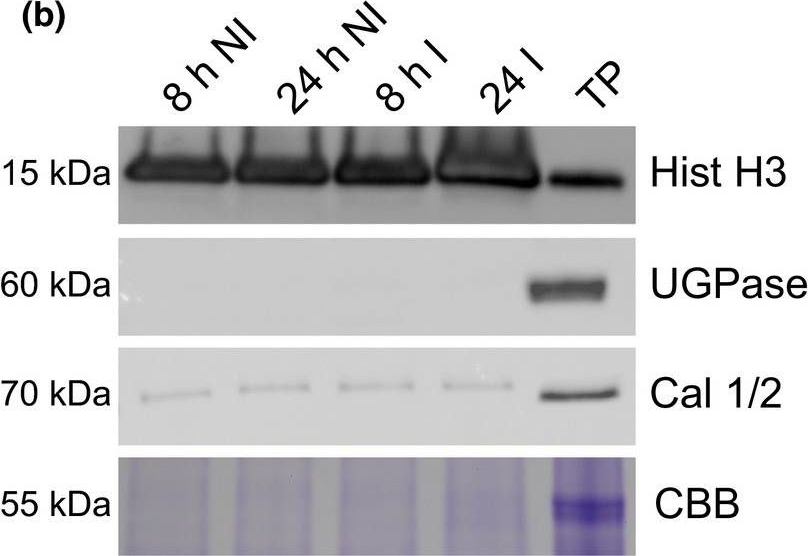
Open PublicationA simple workflow combining nuclear enrichment with quantitative mass spectrometry allows the study of host nuclear processes during infection. (a) Method overview. Detached leaves from 4‐wk‐old tomato plants were spray‐inoculated with Phytophthora capsici zoospores at a concentration of 500 000 spores ml−1, or water as a control (i). Leaves were harvested 8 and 24 h post‐infection and subject to nuclear enrichment (ii). Nuclear protein extracts were generated and fractionated in‐gel and digested with trypsin (iii). Peptide samples were subjected to liquid chromatography−tandem mass spectrometry (LC‐MSMS) analysis and peptide identification and label‐free quantification was carried out using the maxquant and perseus software packages, to identify proteins that were differentially expressed during infection. Nuclear predictions were performed using prediction software on our in‐house Galaxy server as described in the Methods. Subsequent data filtering was completed using the r software package (iv and v). Three independent biological replicates were generated. I, infected samples; NI, noninfected samples (v). (b) Successful enrichment of nuclear proteins demonstrated by western blotting with anti‐histone H3 antibody and subcellular markers. Protein extracts from enriched nuclear samples were compared to a total protein extract (TP) from tomato leaves. Protein concentrations were adjusted for equal loading. Nonnuclear contamination was assessed by probing with anti‐UDP‐glucose pyrophosphorylase (UGPase) antibody (cytoplasm) and calnexin homologue 1/2 antibody (endoplasmic reticulum). Samples were also run on gels and stained with Coomassie brilliant blue (CBB) to assess protein loading and Rubisco abundance. The figure shows the results from a single biological replicate. Blots for all three replicates are provided in Supporting Information Fig. S2.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 30149837
Journal: Elife
Figure Number: 2A
Published Date: 2018-08-28
First Author: Chahtane, H., Nogueira Füller, T., et al.
Impact Factor: 7.448
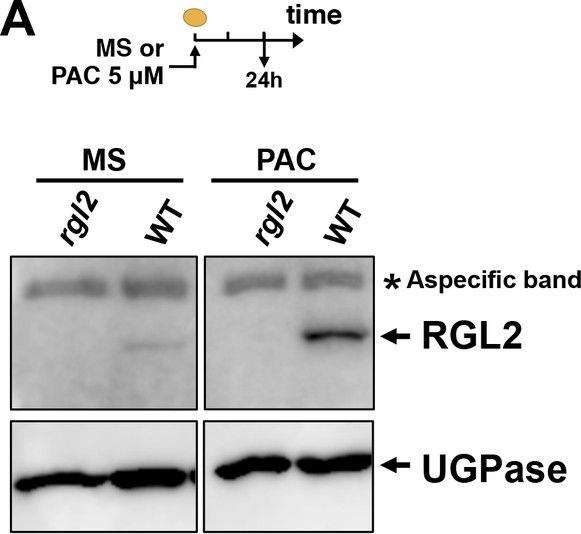
Open PublicationRGL2, GAI, RGA and ABI5 antibodies specificity.Protein gel blot analysis using antibodies to RGL2 (A), ABI5 (B), GAI and RGA (C) as indicated. Protein extracts from Col-0 (WT), rgl2-13 (rgl2), rgl2-SK54 rga-28 (rgl2 rga), rgl2-SK54 rga-28 gai-t6 (rgl2 rga gai) and abi5-3 (abi5) mutant seeds harvested after seed imbibition in presence or absence of PAC (PAC) as indicated. UGPase protein levels are used as a loading control.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 30149837
Journal: Elife
Figure Number: 6D
Published Date: 2018-08-28
First Author: Chahtane, H., Nogueira Füller, T., et al.
Impact Factor: 7.448
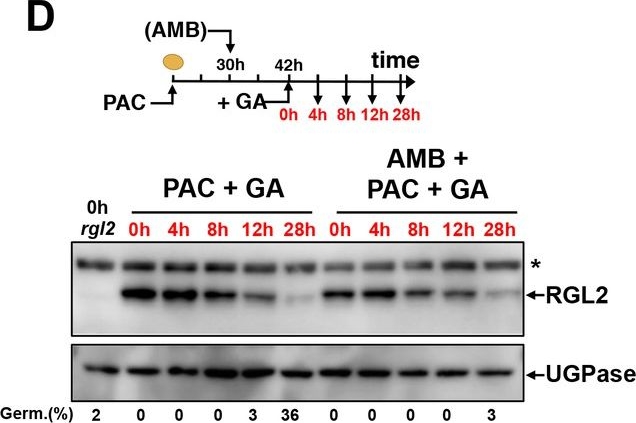
Open PublicationAMB and PAC induce different changes on DELLA protein accumulation.AMB promotes DELLA factor activity to stimulate ABA signaling. (A-C) Protein gel blot analysis using antibodies to RGL2 (A), RGA (B) and GAI (C) as indicated. Protein extracts from WT (Col seeds harvested 24 hr after seed imbibition in absence (MS) or presence of 10 µM PAC (PAC), 5 µM ABA (ABA) or 50 µM AMB (AMB). UGPase protein levels are used as a loading control. (D) AMB does not interfere with GA-dependent RGL2 protein downregulation. WT seeds are imbibed in presence of 5 µM PAC for 30 hr to trigger high RGL2 accumulation. Thereafter, seeds are transferred to germination plates containing 5 µM PAC or containing 5 µM PAC and 50 µM AMB. After 12 hr, 1 µM GA is further added and RGL2 protein levels are followed by protein gel blot analysis over the time points indicated in red. UGPase protein levels are used as a loading control. Germination percentage at each time point is indicated. The asterisk (*) represents and unspecific banc detected by the RGL2 antibody. (E) AMB stimulates DELLA activity to promote ABA-dependent responses. Histograms show RGL2, ABI5, EM1 and NCED6 mRNA accumulation in WT (Col) and Δdella seeds treated as described in A. For each time point, mRNA levels are normalized to mRNA levels in WT seeds sown in absence of AMB. Data represent mean ±standard deviation (three replicates).AMB does not interfere with GA-dependent GAI and RGA protein downregulation.WT seeds are imbibed in presence of 5 µM PAC for 30 hr to trigger high DELLA accumulation. Thereafter, seeds are transferred to germination plates containing 5 µM PAC or containing 5 µM PAC and 50 µM AMB. After 12 hr, 1 µM GA is further added and RGA (A) and GAI (B) protein levels are followed by protein gel blot analysis over the time points indicated in red as indicated. UGPase protein levels are used as a loading control.
- Additional Information
-
Additional information: Cellular [compartment marker] of cytoplasm, UGPse is a cytoplasmic protein Martz et al. (2002)
This product can be sold containing ProClin if requested.Additional information (application): This antibody detectes 1 ng of UGPase in a western blot and reacts with both cytosolic isoforms only which have similar MW of ca. 52 kDa in Arabidopsis thaliana.
For using this antibody on Solanum lycopersicum, 6M urea requires to be included in extraction and loading buffers.
- Background
-
Background: UDP-glucose pyrophosphorylase (UGPase, UDPGP) E.C=2.7.7.9. is a key enzyme of synthesis of sucrose, cellulose and other saccharides. There are two cytoplasmic isoforms of UGPase-A (which share 94 % identity on amino acid level) and one chloroplastic UGPase-B isoform in Arabidopsis thaliana which share ca. 10-11 % of identity (Kleczkowski et al. 2011).
- Product Citations
-
Selected references: Martignago et al (2023) The bZIP transcription factor AREB3 mediates FT signalling and floral transition at the Arabidopsis shoot apical meristem. PLoS Genet. 2023 May 15;19(5):e1010766. doi: 10.1371/journal.pgen.1010766.
Wang et al (2023) Uptake of oomycete RXLR effectors into host cells by clathrin-mediated endocytosis. Plant Cell. 2023 Jun 26;35(7):2504-2526. doi: 10.1093/plcell/koad069.
Boussardon C, Carrie C, Keech O. Comparing plastid proteomes points towards a higher plastidial redox turnover in vascular tissues than in mesophyll cells. J Exp Bot. 2023 Apr 7:erad133. doi: 10.1093/jxb/erad133.
Suanno et al. (2023) Small extracellular vesicles released from germinated kiwi pollen (pollensomes) present characteristics similar to mammalian exosomes and carry a plant homolog of ALIX, Front. Plant Sci., 25 January 2023, Sec. Plant Membrane Traffic and Transport, Volume 14 - 2023.
Vogel et al. (2022) Lipid-mediated activation of plasma membrane-localized deubiquitylating enzymes modulate endosomal trafficking. Nat Commun. 2022;13(1):6897. Published 2022 Nov 12. doi:10.1038/s41467-022-34637-4
Gong et al. (2022). The origin and evolution of a plant resistosome. Plant Cell. 2022 Apr 26;34(5):1600-1620. doi: 10.1093/plcell/koac053. PMID: 35166827; PMCID: PMC9048963.
Hacquard et al. (2022) The Arabidopsis F-box protein FBW2 targets AGO1 for degradation to prevent spurious loading of illegitimate small RNA. Cell Rep. 2022 Apr 12;39(2):110671. doi: 10.1016/j.celrep.2022.110671. PMID: 35417704; PMCID: PMC9035678.
Fang et al. (2022) TANDEM ZINC-FINGER/PLUS3 regulates phytochrome B abundance and signaling to fine-tune hypocotyl growth. Plant Cell. 2022;34(11):4213-4231. doi:10.1093/plcell/koac237
Pan et al. (2021) Post-Golgi Trafficking of Rice Storage Proteins Requires the Small GTPase Rab7 Activation Complex MON1-CCZ1, Plant Physiology, 2021;, kiab175, https://doi.org/10.1093/plphys/kiab175
Li et al. (2021) Isolation and comparative proteomic analysis of mitochondria from the pulp of ripening citrus fruit. Hortic Res. 2021 Feb 1;8(1):31. doi: 10.1038/s41438-021-00470-w. PMID: 33518707; PMCID: PMC7848011.
Li et al. (2020). N-terminal acetylation stabilizes SIGMA FACTOR BINDING PROTEIN 1 involved in salicylic acid-primed cell death. Plant Physiol. 2020 Mar 5. pii: pp.01417.2019. doi: 10.1104/pp.19.01417.
Ren et al. (2020). GPA5 Encodes a Rab5a Effector Required for Post-Golgi Trafficking of Rice Storage Proteins. Plant Cell. 2020 Jan 16. pii: tpc.00863.2019. doi: 10.1105/tpc.19.00863.
Ge et al. (2019). The NIN-like protein 5 (ZmNLP5) transcription factor is involved in modulating the nitrogen response in maize. Plant J. 2019 Dec 2. doi: 10.1111/tpj.14628.
Kim et al. (2019). Polyamine uptake transporter 2 (put2) and decaying seeds enhance phyA-mediated germination by overcoming PIF1 repression of germination. PLoS Genet. 2019 Jul 24;15(7):e1008292. doi: 10.1371/journal.pgen.1008292.
Dogra et al. (2019). Oxidative post-translational modification of EXECUTER1 is required for singlet oxygen sensing in plastids. Nat Commun. 2019 Jun 27;10(1):2834. doi: 10.1038/s41467-019-10760-6.
Pontier et a. (2019). The m6A pathway protects the transcriptome integrity by restricting RNA chimera formation in plants. Life Sci Alliance. 2019 May 29;2(3). pii: e201900393. doi: 10.26508/lsa.201900393.
Jones et al. (2019). Arabidopsis JMJD5/JMJ30 Acts Independently of LUX ARRHYTHMO Within the Plant Circadian Clock to Enable Temperature Compensation. Front. Plant Sci., 01 February 2019 | https://doi.org/10.3389/fpls.2019.00057
Lai et al. (2018). Salicylic acid-independent role of NPR1 is required for protection from proteotoxic stress in the plant endoplasmic reticulum. Proc Natl Acad Sci U S A. 2018 May 29;115(22):E5203-E5212. doi: 10.1073/pnas.1802254115.
Shanmugabalaji et al. (2018). Chloroplast Biogenesis Controlled by DELLA-TOC159 Interaction in Early Plant Development. Curr Biol. 2018 Aug 20;28(16):2616-2623.e5. doi: 10.1016/j.cub.2018.06.006.
Hartmann et al. (2018). Subcellular Compartmentation of Alternatively Spliced Transcripts Defines SERINE/ARGININE-RICH PROTEIN30 Expression. Plant Physiol. 2018 Apr;176(4):2886-2903. doi: 10.1104/pp.17.01260.
Howden et al. (2017), Quantitative analysis of the tomato nuclear proteome during Phytophthora capsici infection unveils regulators of immunity. New Phytol. 2017 Jul;215(1):309-322. doi: 10.1111/nph.14540.
Vincent et al. (2017). A genome-scale analysis of mRNAs targeting to plant mitochondria: upstream AUGs in 5' untranslated regions reduce mitochondrial association. Plant J. 2017 Dec;92(6):1132-1142. doi: 10.1111/tpj.13749.
Nagel et al. (2017). Arabidopsis SH3P2 is an ubiquitin-binding protein that functions together with ESCRT-I and the deubiquitylating enzyme AMSH3. Proc Natl Acad Sci U S A. 2017 Aug 7. pii: 201710866. doi: 10.1073/pnas.1710866114.
Schalk et al. (2017). Small RNA-mediated repair of UV-induced DNA lesions by the DNA DAMAGE-BINDING PROTEIN 2 and ARGONAUTE 1. Proc Natl Acad Sci U S A. 2017 Mar 21. pii: 201618834. doi: 10.1073/pnas.1618834114.
Castellano et al. (2016). A pathogenic long noncoding RNA redesigns the epigenetic landscape of the infected cells by subverting host Histone Deacetylase 6 activity. New Phytol. 2016 Sep;211(4):1311-22. doi: 10.1111/nph.14001. Epub 2016 May 12.
Hsu et al. (2016). Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc Natl Acad Sci U S A. 2016 Oct 21. pii: 201614788.
Liu et al. (2016). iTRAQ-based quantitative proteomic analysis reveals the role of the tonoplast in fruit senescence. J Proteomics. 2016 Sep 2;146:80-9. doi: 10.1016/j.jprot.2016.06.031. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts - Reviews:
-
Manuela Leonardelli | 2020-08-05Very good loading control for arabidopsis stressed plants or mutants affected in chloroplasts functions. I find it more consistent than other controls such as actin/tubulinEirini Kaiserli | 2018-09-14Great antibody - have used it since 2006!
Accessories
AS04 043 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Arabidopsis thaliana, Brassica napus, Macroptilium atropurpureum, Nicotiana benthamiana, Pinus silvestris, Pinus yunanniensis, Oryza sativa, Petunia hybrida cv. Mitchell, Solanum tuberosum, Zea mays | cellular [compartment marker] of cytoplasm