1

Anti-Lhcb1 | LHCII type I chlorophyll a/b-binding protein
AS09 522 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Arabidopsis thaliana, Pinus strobus L.
- Product Info
-
Immunogen: BSA-conjugated synthetic peptide derived from Arabidopsis thaliana At1g29910 (Lhcb1.1), At1g29920 (Lhcb1.2), At1g29930 (Lhcb1.3, most expressed), At2g34430 (Lhcb1.4), and At2g34420 (Lhcb1.5)
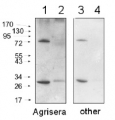
Host: Rabbit Clonality: Polyclonal Purity: Immunogen affinity purified serum in PBS pH 7.4. Format: Lyophilized Quantity: 100 µg Reconstitution: For reconstitution add 100 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Western blot (WB) Recommended dilution: 1 : 2500-1 : 5000 (WB) Expected | apparent MW: 28 | 25 kDa (Arabidopsis thaliana)
- Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Digitaria sanguinalis, Echinochloa crus-galli, Pinus strobus L., Solanum lycopersicum Predicted reactivity: Brassica napus, Camelina sativa, Raphanus sativus, Zea mays
Species of your interest not listed? Contact usNot reactive in: No confirmed exceptions from predicted reactivity are currently known - Application Examples
-
Application examples: 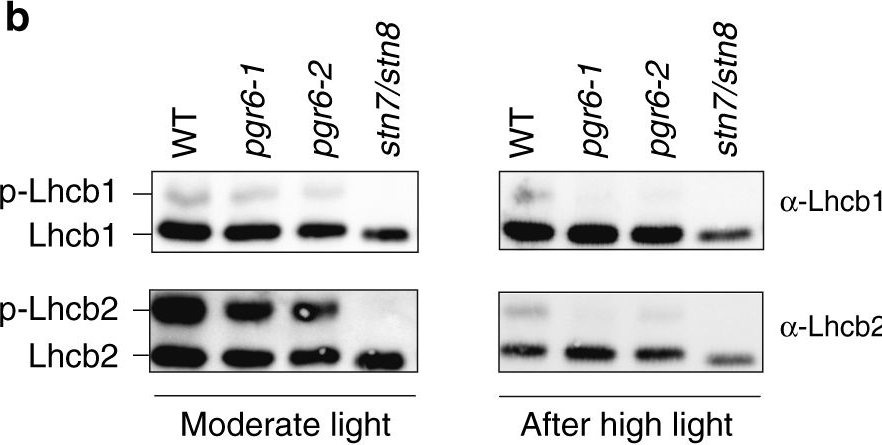
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 31240258
Journal: Commun Biol
Figure Number: 2b
Published Date: 2019-06-27
First Author: Pralon, T., Shanmugabalaji, V., et al.
Impact Factor: None
Open PublicationThylakoid protein phosphorylation and state transitions are disturbed after high light treatment in pgr6 background. a Total protein extracts of 4-week-old wild type (WT), pgr6-1, pgr6?2, sps2 and stn7/stn8 analysed by immunoblotting with anti-phosphothreonine antibody; the principal thylakoid phospho proteins are indicated on the right according to their size. Core photosystem II proteins D1 (PsbA) and D2 (PsbD) are indicated as a single band due to their poor resolution. Actin was used as a loading control. b Lhcb1 and Lhcb2 phosphorylation levels were visualised after separation on Phostag™-pendant acrylamide gels. The upper band corresponds to the phosphorylated form (p-), stn7/stn8 double mutant is a non-phosphorylated control. c Average transient of the variable room temperature chlorophyll fluorescence measured during the transition from red (660nm) supplemented with far-red light (720nm) state 1 to pure red light state 2 (n?=?4 independent pots containing 2–3 plants). The fluorescence curves from pgr6 and sps2 are shifted on the x-axis to allow visualising the FMST1 and FMST2 values. The x-axis time scale refers to the wild-type curve. d Calculated quenching related to state transition (qT?=?(FMST1?–?FMST2)/FM), expressed as the percentage of FM that is dissipated by the state 1 to state 2 transition, of wild type (WT), pgr6?1 and sps2 under moderate light (120?mol?m?2?s?1) (ML) and after 3?h of high light (500?mol?m?2?s?1) (HL). Whiskers and box plot shows the minimum, first quartile, median, average, third quartile and maximum of each dataset (n?=?4 biologically independent samples); p-values are calculated via a two-tailed Student’s t test. e STN7 phosphorylation level visualised after separation on Phostag™-pendant acrylamide gels. The upper band corresponds to the phosphorylated form (p-), a protein sample from stn7/stn8 double mutant was loaded as a control for the antibody specificity. Uncropped images of the membranes displayed in a, b and e are available as Supplementary Fig. 11. Data points for items c, d are available as Supplementary data 2
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 32198392
Journal: Nat Commun
Figure Number: 1B,C
Published Date: 2020-03-20
First Author: Wang, P., Richter, A. S., et al.
Impact Factor: 13.783
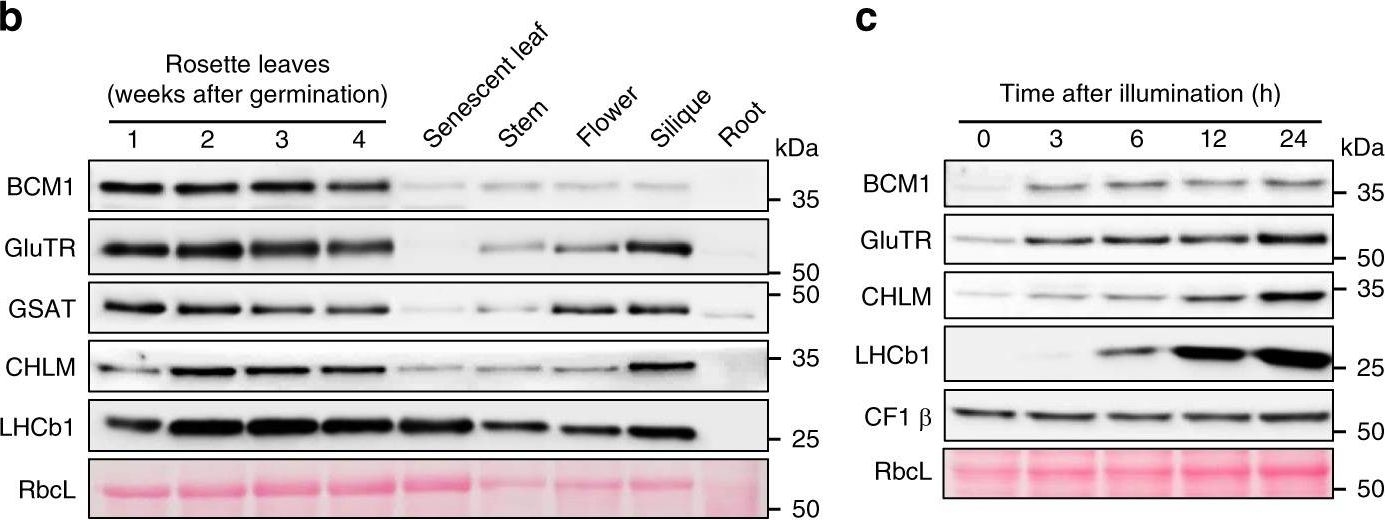
Open PublicationCharacterization of BCM1.a Co-expression analysis of BCM1. BCM1 together with the CBGs, GLK2, the carotenoid biosynthesis gene DEOXYXYLULOSE-5-PHOSPHATE SYNTHASE (DXS), and the gene encoding the chloroplast protein HIGH CHLOROPHYLL FLUORESCENT 107 (HCF107) were hierarchically clustered based on pairwise levels of similarity between their expression profiles, using the gene co-expression database ATTEDII (http://atted.jp/)76. Low distance values indicate high degrees of co-expression. b Accumulation of BCM1, CBEs, and LHCb1 in different organs of Arabidopsis. c Light-induced accumulation of BCM1, CBEs, and LHCb1 in 5-day-old etiolated WT seedlings, following illumination (80??mol photons?m?2?s?1) for 0, 3, 6, 12, and 24?h. d Schematic overview of the domain structure of BCM1. The cTP is shown in gray, while the six predicted TMDs are shown in red. e Subcellular localization of the BCM1-YFP fusion protein. Both BCM1-YFP and cTPBCM1-YFP fluorescence coincides with Chl autofluorescence, confirming chloroplast targeting of BCM1. In the control, YFP itself accumulates in the cytosol and nucleus. Scale bars, 5??m. f Suborganellar localization of BCM1. Chloroplasts from WT seedlings were subfractionated into envelope, stroma, and thylakoid fractions. For comparison, the proteins TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLAST 40 (Tic40), GSAT, and LHC were specifically located in the envelope, stroma, and thylakoid, respectively. g Distribution of BCM1 across the different thylakoid membrane regions. Thylakoid proteins were solubilized with digitonin and fractionated into grana core, grana margins, and stroma lamellae. h Salt washing of thylakoid membranes. The WT thylakoids were sonicated in the presence of 1?M NaCl, 200?mM Na2CO3, 0.1?M NaOH, and 6?M urea on ice for 30?min before centrifugation to separate membrane (P) from soluble (S) fractions. Untreated thylakoids served as the control. LHCb1 and the CF1 ? subunit of ATP synthase, representing an intrinsic membrane protein and a peripheral thylakoid-associated protein, respectively, were used as positive controls for binding affinity to thylakoid membrane fractions. In b, c, f–h, immunoblot analyses were conducted using the indicated antibodies. Ponceau S-stained membrane strips bearing the large subunit of Rubisco (RbcL) or LHC proteins of PSII (LHCII) were used as loading controls.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 32198392
Journal: Nat Commun
Figure Number: 5E
Published Date: 2020-03-20
First Author: Wang, P., Richter, A. S., et al.
Impact Factor: 13.783
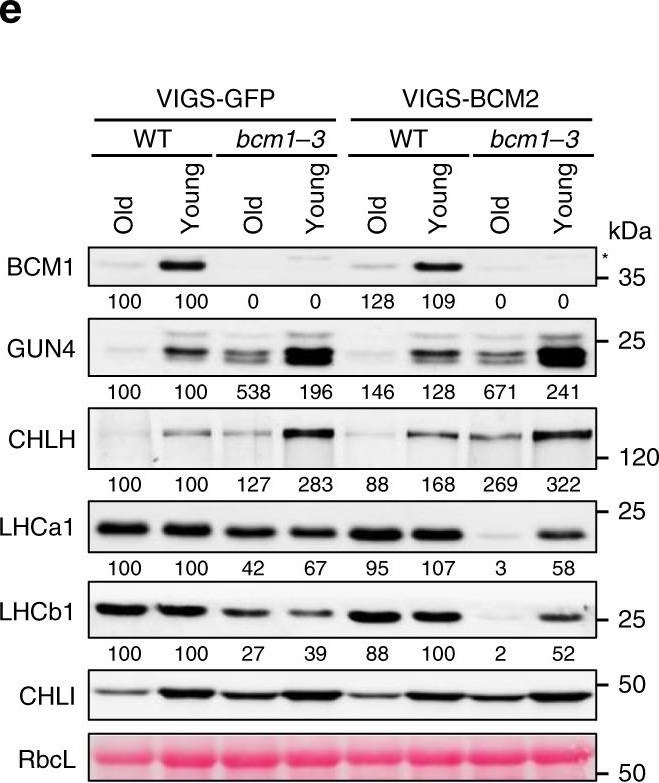
Open PublicationBCM2 plays a conserved role in Chl biosynthesis.a Representative images of 35-day-old VIGS-GFP (as the negative control) and VIGS-BCM2 seedlings, which were infected with the pTRV2-GFP and pTRV2-BCM2 vectors via agroinfiltration in the WT and bcm1-3 background, respectively. The VIGS seedlings were grown under long-day normal light (120??mol photon?m?2?s?1) conditions. The old leaves were indicated by red arrows. b qRT-PCR analysis of BCM1 and BCM2 transcripts in the seedlings shown in a. c Representative images of detached leaves from seedlings shown in a. The old and young mature leaves selected for further analyses were marked by red and white dotted frames, respectively. d Levels of Chl in the old and young leaves shown in c. e Steady-state levels of the indicated proteins in the old and young leaves analyzed in c were determined by immunoblot analysis using the indicated antibodies. f Representative images of 28-day-old WT, bcm1-2, and three independent BCM2-OX/bcm1 transgenic lines overexpressing BCM2 in the bcm1-2 background grown under short-day normal light (120??mol photon?m?2?s?1) conditions. g qRT-PCR analysis of BCM1 and BCM2 transcripts in the seedlings shown in f. Expression levels are presented relative to those in WT seedlings. h, i Levels of Chl (h) and Mg-porphyrin (i) in 18-day-old WT, bcm1-2 and BCM2-OX/bcm1 seedlings grown under short-day normal light (120??mol photon?m?2?s?1) conditions. j Steady-state levels of the indicated proteins in seedlings analyzed in h were determined by immunoblot analysis using the indicated antibodies. In a, c, f, scale bars, 1?cm. In b, g, expression levels are presented relative to those in the VIGS-GFP/WT and WT seedlings, respectively. ND, not detected. In b, d, g–i, error bars represent SD of three biological replicates. Letters above histograms indicate significant differences determined by Tukey’s HSD method (P?<?0.05). In e, j, Ponceau S-stained RbcL was used as the loading control. Numbers below immunoblots represent normalized protein abundances relative to WT seedlings. Asterisks indicate nonspecific signals on the immunoblots.
- Additional Information
-
Additional information: A molecular characterisation of the LHCII proteins can be found in Caffarri et al. (2004) A Look within LHCII: Differential Analysis of the Lhcb1−3 Complexes Building the Major Trimeric Antenna Complex of Higher-Plant Photosynthesis. Biochemistry 43 (29): 9467–9476
Additional information (application): This Lhcb1 antibody is directed specifically against the Arabidopsis Lhcb1 gene products, for those that would prefer higher specific activity over broader specificity offered by Agrisera older Lhcb1 antibody, AS01 004
Protein is processed into mature form (Jansson 1999). - Background
-
Background: The major light-harvesting antenna complex II (LHCII) in photosynthetic eukaryotes is located in the thylakoid membrane of the chloroplast. It is a heterotrimeric complex formed by up to 3 different individual subtypes of chlorophyll a/b-binding proteins: Lhcb1, Lhcb2, and Lhcb3. Lhcb1 is the most abundant chlorophyll a/b-binding protein in eukaryotic phototrophs and often is coded by several nuclear genes.
- Product Citations
-
Selected references: Sakurabata et al. (2024). HASTY-mediated miRNA dynamics modulate nitrogen starvation-induced leaf senescence in Arabidopsis. Nat Commun. 2024 Sep 10;15(1):7913. doi: 10.1038/s41467-024-52339-w.
Bru, Steen, Park, et al. (2022) The major trimeric antenna complexes serve as a site for qH-energy dissipation in plants. J Biol Chem. 2022;298(11):102519. doi:10.1016/j.jbc.2022.102520
Wang et al. (2020). Post-translational coordination of chlorophyll biosynthesis and breakdown by BCMs maintains chlorophyll homeostasis during leaf development. Nat Commun. 2020; 11: 1254.
Pralon et al. (2019). Plastoquinone homoeostasis by Arabidopsis proton gradient regulation 6 is essential for photosynthetic efficiency. Commun Biol. 2019 Jun 20;2:220. doi: 10.1038/s42003-019-0477-4.
Lal et al. (2018). The Receptor-like Cytoplasmic Kinase BIK1 Localizes to the Nucleus and Regulates Defense Hormone Expression during Plant Innate Immunity. Cell Host Microbe. 2018 Apr 11;23(4):485-497.e5. doi: 10.1016/j.chom.2018.03.010.
Tamburino et al. (2017). Chloroplast proteome response to drought stress and recovery in tomato (Solanum lycopersicum L.). BMC Plant Biol. 2017 Feb 10;17(1):40. doi: 10.1186/s12870-017-0971-0.
Fristedt et al. (2017). PSB33 sustains photosystem II D1 protein under fluctuating light conditions. Journal of Experimental Botany doi:10.1093/jxb/erx218.
Hartings et al. (2017). The DnaJ-Like Zinc-Finger Protein HCF222 Is Required for Thylakoid Membrane Biogenesis in Plants. Plant Physiol. 2017 Jul;174(3):1807-1824. doi: 10.1104/pp.17.00401.
Correa-Galvis et al. (2016). PsbS interactions involved in the activation of energy dissipation in Arabidopsis. Nature Plants 2, Article number: 15225 (2016) doi:10.1038/nplants.2015.225
Longoni et al. (2015). Phosphorylation of the Lhcb2 isoform of Light Harvesting Complex II is central to state transitions. Plant Physiol. 2015 Oct 5. pii: pp.01498.2015.
Wientjes et al (2013). LHCII is an antenna of both photosystems after long-term acclimation. BBA, Jan 6. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
Oxygenic photosynthesis poster by prof. Govindjee and Dr. Shevela
Z-scheme of photosynthetic electron transport by prof. Govindjee and Dr. Björn and Dr. Shevela - Reviews:
-
Pierrick | 2020-11-16This antibody gives a clear single band with Arabidopsis leaf protein extracts (from leaf punch). I blocked the membranes with TBST + 3% milk for 1h at RT. The primary antibodies were diluted in TBST + 3% milk 1:5000 and incubated with the membranes for 1 h at RT with agitation. I�m reusing the diluted antibodies for up to 5 to 10 western blots without any significant loss of signal (Diluted antibodies are stored at -20�C). The secondary antibodies were diluted in TBST + 3% milk 1:10000 and incubated with the membranes for 1 h at RT with agitation.Tanja G�bel | 2012-09-25I�m using the antibodies for the immunodetection on western blots with the fluorescent based licor odyssey system (with the IRDye secondary antibodies Donkey anti rabbit IgG 800CW and 680 (Licor)). I�m analysing whole protein or chloroplasts protein extracts optained from Arabidopsis thaliana. Proteins were either precipitated with TCA/Aceton or directly solubilized with NP-40 in a tris-buffer. The only problem with the unprecipitated samples is that one gets a very strong backround in the 680nm channel due to the chlorophyll-fluorescence. The primary antibodies were diluted in TBS with 0,02% Sodiumazid according to the recommanded dilution (for ECL) in the data sheet and the membranes were incubated for 1 h at RT with agitation. The membranes are blocked with BSA in TBS and washed with TBS-T. I�m reusing the diluted antibodies for up to 5 to 10 western blots without any significant loss of signal (diuluted antibodies are stored at 4�C). The signals I�m getting with the antibodies are very clear and hieghly specific, with only low (anti-Lhcb1 and 2) or nearly no (anti-RbcL and RA) unspecific binding. Usually, I�m loading 10 �g protein and seperate them on 10% Mini-gels. So far I had no problems with Agrisera-antibodies.